Hi everyone,
I’ve done a lot of deep learning work for chemical synthesis using SMILES representations and transformer models. These have worked quite well, but I want to work with graph-based networks as well to compare performance. I haven’t quite wrapped my head around GCNN/GATs yet, and I wanted to ask some clarifying questions.
Source vectors for attention
As I understand it, attention within a node is applied over all feature vectors sent to that node from direct neighbors. If this is correct, is this technique restricted to direct neighbors, or can a node gather and attend to all features in a graph?
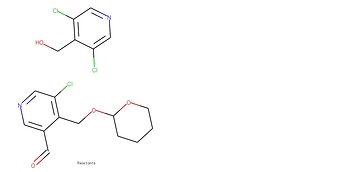
Going off that, does the GAT structure allow a node to attend to a different disconnected graph? The specific example I’m looking at is the case of two chemical reactants, like these:
In graph form, these two molecules are separate graphs. Is it possible for nodes on one molecule graph to attend to or otherwise consider nodes on the other molecule graph? (For reference, the ability for one chemical token to attend to all tokens in the reaction is a major reason why transformers work extremely well on this task).
One thought I had when dealing with this was to run a transformer type attention layer over the entire feature vector after a message passing step, but this gets into my next question.
Handling batching when multiple examples contain disconnected graphs
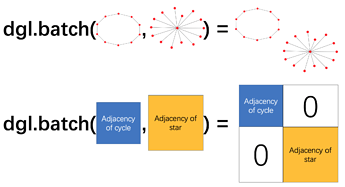
Going off the documentation, batching graphs is handled by merging graphs like so:
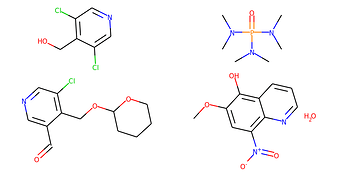
Batching implies that each input example is a single discrete graph. Now consider two chemical examples:
Here we have two examples. Two reactant sets. Each reactant set contains two separate molecule graphs. If I packaged both examples into a graph batch, it would look the same as four separate examples. How do I represent this kind of data in batched form that preserves the notion that sometimes separate graphs belong to the same example?
Thanks