Thanks for the reply!
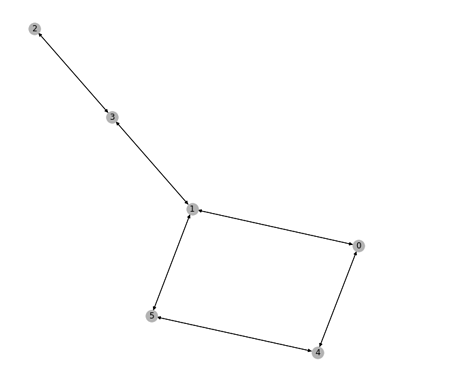
Sorry but I still dont get it though. I’ve simplified the Graph in the above example to be:
Can you please explain what do you mean by “last block” here? As seen below, each step here has just 1 block.
Data loader creation code:
# edges to compute output
train_eids = th.tensor([0, 1])
print(print("edges to compute output, src, dst are: ", G.find_edges(train_eids)))
fanouts = [1] # List of neighbors to sample for each GNN layer, let's say just one layer and one neighbor.
sampler = dgl.dataloading.MultiLayerNeighborSampler(fanouts)
negative_sampler = NegativeSampler(G, 2) # 2 negative samples per positive
# define the dataloader:
dataloader = dgl.dataloading.EdgeDataLoader(
G,
train_eids,
sampler,
exclude=None,
negative_sampler=negative_sampler,
batch_size=1,
shuffle=True,
drop_last=False,
pin_memory=True)
Printing the EdgeDataLoader iteration output:
for step, (input_nodes, pos_graph, neg_graph, blocks) in enumerate(dataloader):
assert sum(th.eq(pos_graph.nodes(), neg_graph.nodes())) == \
neg_graph.number_of_nodes() == \
pos_graph.number_of_nodes()
print("************ step-{} **********".format(step))
print("input_nodes: ", input_nodes)
print("pos_graph {} edges: ".format(pos_graph.number_of_edges()), pos_graph.edges())
print("pos_graph {} nodes: ".format(pos_graph.number_of_nodes()), pos_graph.nodes())
print("pos_graph {} original edges: ".format(pos_graph.number_of_edges()), pos_graph.edata[dgl.EID])
print("pos_graph {} original nodes: ".format(pos_graph.number_of_nodes()), pos_graph.ndata[dgl.NID])
# neg_graph edges number == number defined in NegativeSampler()
print("neg_graph {} edges: ".format(neg_graph.number_of_edges()), neg_graph.edges())
print("neg_graph {} nodes: ".format(neg_graph.number_of_nodes()), neg_graph.nodes())
for b in blocks:
print("\tblock nodes number: ", b.number_of_nodes())
print("\tblock nodes: ", b.nodes("_U"))
print("\tblock edges: ", b.edges("uv"))
And get this output:
************ step-0 **********
input_nodes: tensor([1, 0, 5, 4])
pos_graph 1 edges: (tensor([0]), tensor([1]))
pos_graph 3 nodes: tensor([0, 1, 2])
pos_graph 1 original edges: tensor([0])
pos_graph 3 original nodes: tensor([1, 0, 5])
neg_graph 2 edges: (tensor([0, 0]), tensor([1, 2]))
neg_graph 3 nodes: tensor([0, 1, 2])
block nodes number: 7
block nodes: tensor([0, 1, 2, 3])
block edges: (tensor([1, 3, 3]), tensor([0, 1, 2]))
************ step-1 **********
input_nodes: tensor([2, 3, 5, 1, 4, 0])
pos_graph 1 edges: (tensor([0]), tensor([1]))
pos_graph 4 nodes: tensor([0, 1, 2, 3])
pos_graph 1 original edges: tensor([1])
pos_graph 4 original nodes: tensor([2, 3, 5, 1])
neg_graph 2 edges: (tensor([0, 0]), tensor([2, 3]))
neg_graph 4 nodes: tensor([0, 1, 2, 3])
block nodes number: 10
block nodes: tensor([0, 1, 2, 3, 4, 5])
block edges: (tensor([1, 3, 4, 5]), tensor([0, 1, 2, 3]))
Looking at the first step -
It seems the edges in the block do not correlate to either pos or neg graph. Why is that?