Q1:
The h's are node features in trees. The tree representations can be obtained by some function taking into node features, edge features, tree structure, etc. Since you are interested in the learned representations, you will only need to save the results for the last epoch.
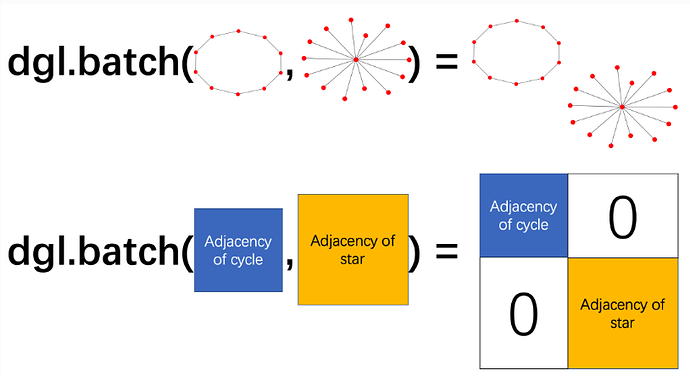
In dgl, when we batch a collection of graphs/trees, we are essentially merging them into a giant graph with several isolated connected components:
Then in the example you gave this means we will have a graph consisting of 5 isolated connected components, with a total number of 273 nodes. The nodes in the new giant graph are re-indexed as consecutive integers starting from 0. The example below might be helpful:
import dgl
import torch
g_list = []
# Create a list of graphs respectively having 1, 2, 3 nodes
for i in range(3):
g = dgl.DGLGraph()
g.add_nodes(i + 1)
# The node features for the 3 graphs are separately
# [[0]], [[1], [2]], [[3], [4], [5]]
start = sum(range(i + 1))
g.ndata['h'] = torch.range(start, start + i).view(-1, 1)
g_list.append(g)
# Merge the list of graphs into one giant graph
bg = dgl.batch(g_list)
print(bg.ndata['h']) # [[0], [1], [2], [3], [4], [5]]
# You can also query the number of nodes in each original graph
print(bg.batch_num_nodes) # [1, 2, 3]
With the batched graph, the node features in all original trees can be updated in parallel. Note that the update happening in batched graph will not come into effect on the original graphs. You can convert the merged large graph back into the small graphs and replace the original g_list.
bg.ndata['h'] += 1
print(bg.ndata['h']) # [[1.], [2.], [3.], [4.], [5.], [6.]]
print(g_list[2].ndata['h'] # [[3.], [4.], [5.]]
# Replace the original list of graphs
g_list = dgl.unbatch(bg)
for i in range(3):
print(g_list[i].ndata['h']) # [[1.]], [[2.], [3.]], [[4.], [5.], [6.]]
Q2:
You can always store different node features and use these node features to compute some objective function. If you want to use some your own dataset, you can write a dataset class yourself. The source code for SST is here for your reference.
For the last part,
Additionally, my ultimate goal is to do something similar to the sentence classification goal, but instead of the supervision being done by comparing the root of two trees, a few nodes within a given tree should (or should not) have the same label as other sporadic nodes within another tree.
my very rough feeling is that you want to compute the pairwise (dis-)similarity between node embeddings from different trees. My feeling is that it might be more straightforward to write some PyTorch code yourself.