Hi, a newbie question here. I’m trying to write a syntactic parser using the DGL library but I’m struggling with understanding the model training loop. In a link prediction task, the available tutorials and documentation only show looping through one large graph. Given my task at hand, though, I want to treat each sentence in a corpus as a graph. Can DGL handle training on multiple small graphs to predict edges? If so, should I use a batch variable instead of the single graph variable at every step of the pipeline? Thanks in advance.
One issue you may hit when apply link prediction on the batched graph: negative sampling. you need to have your own negative sampling to avoid sample edges whose src node and dst node belongs to different original graph. here’s a reference: dgl.sampling.global_uniform_negative_sampling — DGL 0.8.0post1 documentation
Let’s consider an example of two graphs below.
import dgl
g1 = dgl.rand_graph(num_nodes=10, num_edges=30)
g2 = dgl.rand_graph(num_nodes=15, num_edges=50)
# Batch the two graphs
bg = dgl.batch([g1, g2])
You can use the batched graph bg for message passing over multiple graphs in parallel. When it comes to sampling negative edges, having multiple graphs introduces additional complexity as you do not want negative edges between nodes in different graphs. In this particular example, we want to sample negative edges among nodes in g1 and among nodes in g2, but not between nodes in g1 and g2. So far, DGL does not have built-in support for this case. The code snippet below provides an example for negative sampling over multiple graphs.
import torch
num_neg_edges_g1 = 6
num_neg_edges_g2 = 10
num_neg_edges_bg = num_neg_edges_g1 + num_neg_edges_g2
# Get normalized relative index
neg_norm_src = torch.rand(num_neg_edges_bg)
neg_norm_dst = torch.rand(num_neg_edges_bg)
# Get start node ID for each negative edge
batch_num_nodes = bg.batch_num_nodes()
batch_num_nodes = torch.cat([torch.zeros(1), batch_num_nodes])
batch_num_nodes_cumsum = torch.cumsum(batch_num_nodes, dim=0)
neg_start = torch.cat([
torch.zeros(num_neg_edges_g1).fill_(batch_num_nodes_cumsum[0]),
torch.zeros(num_neg_edges_g2).fill_(batch_num_nodes_cumsum[1])])
neg_full_range = torch.cat([
torch.zeros(num_neg_edges_g1).fill_(batch_num_nodes[1]),
torch.zeros(num_neg_edges_g2).fill_(batch_num_nodes[2])])
neg_src = (neg_start + neg_norm_src * neg_full_range).long()
neg_dst = (neg_start + neg_norm_dst * neg_full_range).long()
Thank you, this is very helpful! However one thing I’m not sure about are the initial num_neg_edges values (6 and 10 here). Are they arbitrary? Or is there a specific reason for them to be a result of a num_edges/5 division?
They are arbitrary and you can control them by hyperparameter(s). For example, you can use a pre-specified number of negative edges for all graphs or a pre-specified ratio of negative edges for all graphs.
Thanks!
My other another conceptual struggle with a batch dataset is about the train/test split. In the Link Prediction tutorial (the snippet below), the data of a large single graph is split before the negative edges are obtained. However, if the same procedure is applied to a batch, will the integrity of each graph in the batch be preserved in the training? Should the data be split instead by graph IDs rather than by edge IDs, with negative edges obtained subsequently from train and test data?
# Split edge set for training and testing
u, v = batch_g.edges()
eids = np.arange(batch_g.number_of_edges())
eids = np.random.permutation(eids)
test_size = int(len(eids) * 0.1)
train_size = batch_g.number_of_edges() - test_size
test_positive_u, test_positive_v = u[eids[:test_size]], v[eids[:test_size]]
train_positive_u, train_positive_v = u[eids[test_size:]], v[eids[test_size:]]
# Find all negative edges and split them for training and testing:
adj = sp.coo_matrix((np.ones(len(u)), (u.numpy(), v.numpy())))
adj_neg = 1 - adj.todense() - np.eye(batch_g.number_of_nodes())
neg_u, neg_v = np.where(adj_neg != 0)
neg_eids = np.random.choice(len(neg_u), batch_g.number_of_edges())
test_negative_u, test_negative_v = neg_u[neg_eids[:test_size]], neg_v[neg_eids[:test_size]]
train_negative_u, train_negative_v = neg_u[neg_eids[test_size:]], neg_v[neg_eids[test_size:]]
It might make more sense to perform edge split for individual graphs, i.e., before batching them.
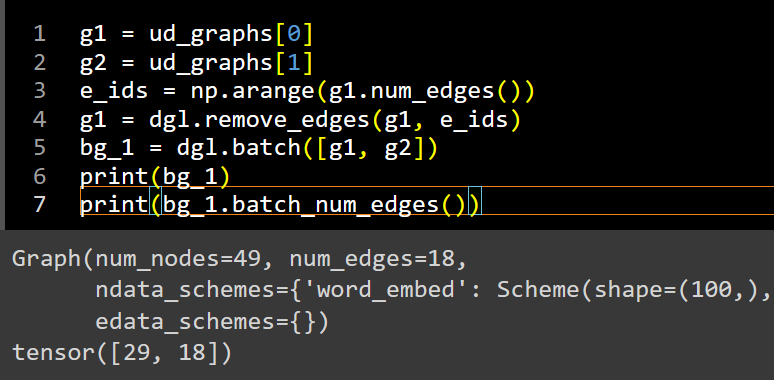
This went well but now I’m facing another issue: after batching graphs with some of their edges removed for training (with remove_edges()), the total num_edges of the batch correctly shows a reduced count, whereas batch_num_edges lists edge counts equal to those in the graphs before edge removal (as if no edges were removed). Where is this discrepancy coming from?
couldn’t replicate the issue.
Maybe could you run again and then post the full code again including your graphs creation too?
please post as a Preformatted text (ctrl+e) instead of image?
You’re right, the issue doesn’t replicate, except for when I use graphs from my dataset. The snippet below shows 0 edges for g1 in both queries (even when I added ndata to resemble my dataset):
g1 = dgl.rand_graph(num_nodes=10, num_edges=30)
g2 = dgl.rand_graph(num_nodes=15, num_edges=50)
e_ids = np.arange(g1.num_edges())
g1 = dgl.remove_edges(g1, e_ids)
batched = dgl.batch([g1, g2])
print(batched)
print(batched.batch_num_edges())
Not sure what the difference could be in my graphs and why the edge information is only partially carried into the batch.
try looking at your g1 before and after edge removal.
check the original g1 e_ids too
Hi, I tried to download this module but got this error when I used it:
name '_CAPI_DGLGlobalUniformNegativeSampling' is not defined
Any idea how to work around that? Thanks!
I tried with dgl-cu102 0.8.1 and it works well.
How did you download and which version are you using?
This pip install ends up with an error:
Could not find a version that satisfies the requirement 0.8.1 (from versions: none)
No matching distribution found for 0.8.1
Perhaps it’s because I’m on Colab? But the same error appears whenever I’m trying to install any package from a cloned github repo (following the instructions here). I checked and all the requirements are satisfied so I’m probably making some rookie mistake somewhere…
any reason for building DGL on your own? pls find appropriate cmd and install pre-built DGL in Deep Graph Library
This topic was automatically closed 30 days after the last reply. New replies are no longer allowed.