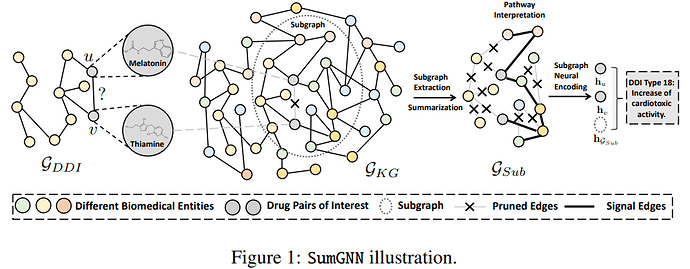
Hi @BarclayII, I wrote the functions below. I am mostly posting it so other people can use it if they need it, but If you have the time, would you mind sharing if you think that there is a faster/better way of doing it? But only if you think it is a quick thing to do, otherwise, I am already thankful for your initial help!
from collections import defaultdict
import numpy as np
import matplotlib.pyplot as plt
from datetime import datetime
import networkx as nx
import dgl
import torch
node_range = 5
number_edges = 20
hg = dgl.heterograph({
('user', 'plays', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
('store', 'sells', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
})
def get_subgraph_from_heterograph(hg, seed_node_1, seed_node_2, k_hops, fanout, print_time):
"""
This function will create a subgraph around an edge pair, given the original heterogenoeus graph.
ex:
node_range = 5
number_edges = 20
hg = dgl.heterograph({
('user', 'plays', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
('store', 'sells', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
})
It will output:
- All the nodes in the subgraph in the format: dict(node_type:node_id)
ex: {'store': [0, 1, 2, 4], 'game': [0, 1, 2, 3, 4], 'user': [0, 1, 2, 3, 4]}
- The distances of all the nodes to each seed_node in the format: dict(node_type: tensor of shape (n_nodes,2))
All nodes have distace -1, and if they are less than k_hops away, then their distances are 0 to k_hops.
ex: defaultdict(dict,
{'game': tensor([[ 2., 0.],
[ 2., 1.],
[-1., 1.],
[ 2., 1.],
[ 0., 1.]]),
'store': tensor([[-1., 0.],
[-1., 2.],
[ 1., 1.],
[ 1., -1.],
[-1., 2.]]),
'user': tensor([[ 1., 2.],
[ 0., 2.],
[ 1., 0.],
[-1., 0.],
[-1., 0.]])})
Parameters
----------
hg : dgl.Heterograph.
It assumes that all IDs are continuous from 0 to n_ID. If it is not the case,
distances will raise an error, because the node ID is used as index.
seed_node_1 : dict(node_type: [node_id])
One of the node in the center pair
seed_node_2 : dict(node_type: [node_id])
The other node in the center pair
k_hops : int
Number of hops around the center edge
fanout : int
Number of node, per edge type, to get at each hop. fanout = -1 will get all connected nodes
print_time: bool True/False
Prints the time it took to create the subgraph
"""
t1 = datetime.now()
distances = defaultdict(dict)
for node_type in hg.ntypes:
distances[node_type] = -torch.ones(hg.num_nodes(node_type), 2)
idx = 0
nodes_subgraph_1 = defaultdict(set)
nodes_subgraph_2 = defaultdict(set)
for nodes_subgraph, seed_node in zip([nodes_subgraph_1, nodes_subgraph_2], [seed_node_1, seed_node_2]):
subgraph_in = dgl.sampling.sample_neighbors(hg, nodes = seed_node, fanout = fanout, edge_dir ='in')
subgraph_out = dgl.sampling.sample_neighbors(hg, nodes = seed_node, fanout = fanout, edge_dir ='out')
for node_type, node_ids in seed_node.items():
distances[node_type][node_ids, idx] = 0
nodes_subgraph[node_type].update(node_ids)
for k_hop in range(k_hops):
subgraphs = [subgraph_in, subgraph_out]
new_adj_nodes = defaultdict(set)
for subgraph in subgraphs:
for node_type_1, edge_type, node_type_2 in subgraph.canonical_etypes:
nodes_id_1, nodes_id_2 = subgraph.all_edges(etype = edge_type)
new_adj_nodes[node_type_1].update(set(nodes_id_1.numpy()).difference(nodes_subgraph[node_type_1]))
new_adj_nodes[node_type_2].update(set(nodes_id_2.numpy()).difference(nodes_subgraph[node_type_2]))
nodes_subgraph[node_type_1].update(new_adj_nodes[node_type_1])
nodes_subgraph[node_type_2].update(new_adj_nodes[node_type_2])
new_adj_nodes = {key:list(value) for key, value in new_adj_nodes.items()}
subgraph_in = dgl.sampling.sample_neighbors(hg, nodes = dict(new_adj_nodes), fanout = fanout, edge_dir ='in')
subgraph_out = dgl.sampling.sample_neighbors(hg, nodes = dict(new_adj_nodes), fanout = fanout, edge_dir ='out')
for node_type, node_ids in new_adj_nodes.items():
distances[node_type][node_ids, idx] = k_hop + 1
idx += 1
#merge nodes_subgraph_1 and nodes_subgraph_2
nodes_subgraph = dict()
for node_type in nodes_subgraph_2.keys():
nodes_subgraph[node_type] = list(nodes_subgraph_1[node_type].union(nodes_subgraph_2[node_type]))
t2 = datetime.now()
if print_time:
print('time:', t2 - t1)
return nodes_subgraph, distances
def draw_graph(subgraph):
"""
Draws a homogeneous graph
"""
g_nx = homo_g.to_networkx(node_attrs=['_TYPE'], edge_attrs=['_TYPE'])
node_attr_dict = nx.get_node_attributes(g_nx, '_TYPE')
node_color = torch.tensor(list(node_attr_dict.values()))
# pos = nx.random_layout(g_nx)
pos = nx.spring_layout(g_nx)
plt.figure(figsize = (10,10))
nx.draw(g_nx,
pos,
node_color = node_color,
with_labels=True,
)
def get_subgraph_from_homograph(g, seed_node_1, seed_node_2, k_hops, print_time):
"""
This function will create a subgraph around an edge pair, given the a homogeneous graph.
ex:
node_range = 5
number_edges = 20
hg = dgl.heterograph({
('user', 'plays', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
('store', 'sells', 'game'): (torch.randint(0, node_range, (number_edges, 1)).reshape(-1), torch.randint(0, node_range, (number_edges, 1)).reshape(-1)),
})
g = dgl.to_homogeneous(hg)
It will output:
- All the nodes in the subgraph in the format: [node_ids]
ex: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14]
- The distances of all the nodes to each seed_node in the format: tensor of shape (n_nodes,2)
All nodes have distace -1, and if they are less than k_hops away, then their distances are 0 to k_hops.
ex: tensor([[0., 2.],
[2., 1.],
[2., 2.],
[2., 2.],
[2., 1.],
[2., 1.],
[1., 2.],
[2., 3.],
[1., 1.],
[2., 1.],
[2., 0.],
[1., 2.],
[1., 2.],
[1., 2.],
[1., 2.]])
Parameters
----------
g : dgl.graph
It assumes that all IDs are continuous from 0 to n_ID. If it is not the case,
distances will raise an error, because the node ID is used as index.
seed_node_1 : [node_id]
One of the node in the center pair
seed_node_2 : [node_id]
The other node in the center pair
k_hops : int
Number of hops around the center edge
print_time: bool True/False
Prints the time it took to create the subgraph
"""
t1 = datetime.now()
distances = -torch.ones(g.num_nodes(), 2)
idx = 0
nodes_subgraph_1 = set()
nodes_subgraph_2 = set()
for nodes_subgraph, seed_node in zip([nodes_subgraph_1, nodes_subgraph_2], [seed_node_1, seed_node_2]):
nodes_subgraph.update(seed_node)
subgraph_in = g.in_edges(seed_node)
subgraph_out = g.out_edges(seed_node)
distances[seed_node, idx] = 0
for k_hop in range(k_hops):
new_adj_nodes = set()
for nodes_id_1, nodes_id_2 in [subgraph_in, subgraph_out]:
nodes_id = set(nodes_id_1.numpy()).union(set(nodes_id_2.numpy()))
new_adj_nodes.update(nodes_id.difference(nodes_subgraph))
nodes_subgraph.update(new_adj_nodes)
new_adj_nodes = list(new_adj_nodes)
subgraph_in = g.in_edges(new_adj_nodes)
subgraph_out = g.out_edges(new_adj_nodes)
distances[new_adj_nodes, idx] = k_hop + 1
idx += 1
nodes_subgraph = list(nodes_subgraph_1.union(nodes_subgraph_2))
t2 = datetime.now()
if print_time:
print('time:', t2 - t1)
return nodes_subgraph, distances
########## HETEROGENEOUS ##########
seed_node_1 = {'game':[0]}
seed_node_2 = {'user':[0]}
print_time = True
k_hops = 3
fanout = -1
nodes_subgraph, hetero_distances = get_subgraph_from_heterograph(hg, seed_node_1, seed_node_2, k_hops, fanout, print_time)
for node_type in hg.ntypes:
hg.nodes[node_type].data['center_dist'] = hetero_distances[node_type]
subgraph = hg.subgraph(nodes_subgraph)
homo_g = dgl.to_homogeneous(subgraph)
draw_graph(homo_g)
########## HOMOGENEOUS ##########
seed_node_1 = [0]
seed_node_2 = [hg.num_nodes('game')+hg.num_nodes('store')]
k_hops = 3
print_time = True
g = dgl.to_homogeneous(hg)
nodes_subgraph, distances = get_subgraph_from_homograph(g, seed_node_1, seed_node_2, k_hops, print_time)
g.ndata['center_dist'] = distances
subgraph = g.subgraph(nodes_subgraph)
draw_graph(subgraph)