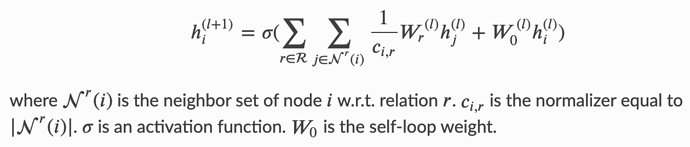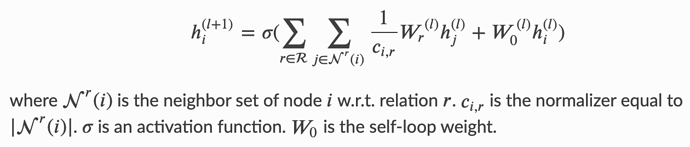Hi Minjie,
thank you for your reply and the further explanation! I am currently working with the example you provided and that seems to work!
I have one more question, which is slightly related, so maybe you can help me. I think I need to shortly introduce my setup for this to make sense (apologies for the long question):
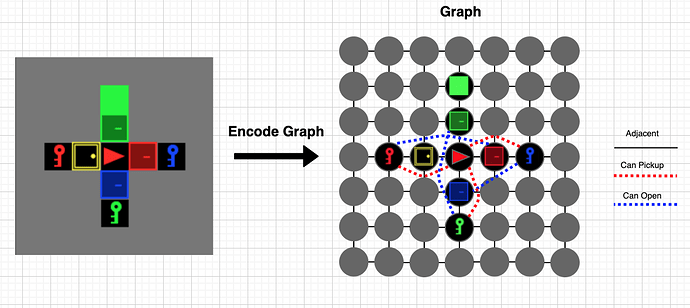
I am using DGL for reinforcement learning on a grid world environment. Each observation is a description of the entire grid, which tells the agent what is present at each grid cell. These observations are encoded as a graph, which will hopefully enable me to add some relational reasoning between elements in the observation. I would like to work with R-GCN and the observations are encoded into a graph as follows:
- Each grid cell is a node in the graph
- There is a ’next to’ relation/edge between all adjacent grid cells, these edges are always the same for each observation as the actual size of the grid does not change
- There are two more types of edges which indicate specific relations between certain items in the grid world. In my case these are edges between keys and the corresponding doors they open and edges between the agent and these keys. As the positions of these objects change, these edges will also have to change for each observation.
A rough sketch of this idea:
The question:
Is there an efficient way to create these graphs and preferably even an efficient way to create a batch of these graphs? Currently I am sending a mini batch of observations - which are edgelists for each relation as I cannot actually store graphs as observations due to the observations spaces of Gym - to my forward pass. In the forward pass I make all the the graphs from these observations and then batch them, but this is very slow.
One option I was thinking of was to actually do use the homogeneous graphs and then I could add/remove the two types of relational edges - ‘Can Pickup’ and ‘Can Open’ - which change per observation? Another option could maybe be to make a fully connected graph for the 2 relations that change per observation and then adapt the weights of these edges to select some of them based on the observation?
If you have an idea on how to do this more efficient, it would be highly appreciated!!
Kind regards,
Erik