Hi,
I had implemented a modification from PinSAGE for my Master’s thesis, but the model is not converting to any result. I would like to know if I’m making some mistake in my DGL or if I need to check other aspects of my formulation.
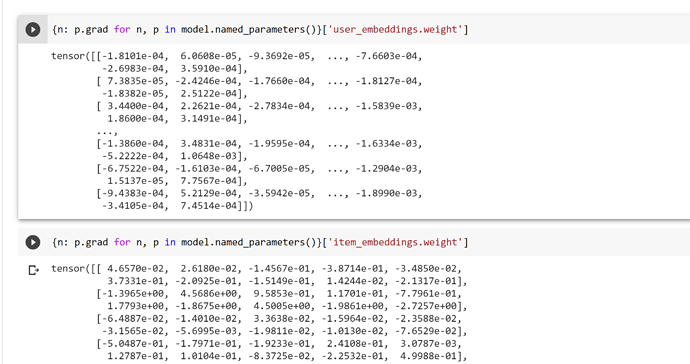
I’m suspecting that for some reason my implementation is using the initial feature vector for the nodes in the convolutions, and not the learned embeddings.
I used the following code:
Scorer
class BAMGScorer(nn.Module):
def __init__(self, num_users, num_items, user_hidden_dim, item_hidden_dim, final_hidden_dim, num_layers=1):
super().__init__()
self.layers = nn.ModuleList([
BAMGLayer(user_hidden_dim, item_hidden_dim) for _ in range(num_layers)])
self.num_users = num_users
self.num_items = num_items
# Node-specific learnable embeddings
self.user_embeddings = nn.Embedding(self.num_users, user_hidden_dim)
self.item_embeddings = nn.Embedding(self.num_items, item_hidden_dim)
def get_representation(self):
return self.user_embeddings, self.item_embeddings
def forward(self, blocks):
user_embeddings = self.user_embeddings(blocks[0].srcnodes['user'].data[dgl.NID])
item_embeddings = self.item_embeddings(blocks[0].srcnodes['item'].data[dgl.NID])
for block, layer in zip(blocks, self.layers):
user_embeddings, item_embeddings = layer(block, user_embeddings, item_embeddings)
return user_embeddings, item_embeddings
def compute_score(self, pair_graph, user_embeddings, item_embeddings):
with pair_graph.local_scope():
pair_graph.nodes['user'].data['h'] = user_embeddings
pair_graph.nodes['item'].data['h'] = item_embeddings
ngh_scorer = NeighborhoodScore(pair_graph)
scores = ngh_scorer.get_similarity_scores()
return scores
Layer
class BAMGLayer(nn.Module):
def __init__(self, user_hidden_dim, item_hidden_dim):
super().__init__()
self.heteroconv = dglnn.HeteroGraphConv(
{'watched': BAMGConv(item_hidden_dim, user_hidden_dim), 'watched-by': BAMGConv(user_hidden_dim, item_hidden_dim)})
def forward(self, block, input_user_features, input_item_features):
with block.local_scope():
h_user = input_user_features
h_item = input_item_features
src_features = {'user': h_user, 'item': h_item}
dst_features = {'user': h_user[:block.number_of_dst_nodes('user')], 'item': h_item[:block.number_of_dst_nodes('item')]}
result = self.heteroconv(block, (src_features, dst_features))
return result['user'], result['item']
Convolution
class BAMGConv(nn.Module):
def __init__(self, src_dim, dest_dim):
super().__init__()
self.linear_src = nn.Linear(in_features=src_dim, out_features=dest_dim, bias=True)
self.linear_dst = nn.Linear(in_features=dest_dim, out_features=dest_dim, bias=True)
def compute_message(self, edges):
affinity = edges.data['weight'] / torch.sum(edges.data['weight'])
return affinity
def forward(self, graph, node_features):
with graph.local_scope():
src_features, dst_features = node_features
graph.srcdata['h'] = src_features
graph.dstdata['h'] = dst_features
graph.apply_edges(lambda edges: {'a': self.compute_message(edges)})
graph.update_all(message_func=fn.u_mul_e('h', 'a', 'h_ngh'),
reduce_func=fn.sum('h_ngh', 'neighbors_avg'))
result = F.relu( torch.add( self.linear_src(graph.dstdata['h']), self.linear_dst(graph.dstdata['neighbors_avg']) ) )
return result
Loss
def compute_margin_loss(scores, margin=0.1):
loss = 0
for score in scores:
loss += (- score['positive_score'] + score['negative_score'] + margin).clamp(min=0)
return loss
Training Loop
NUM_LAYERS = 1
user_hidden_dim = 7
item_hidden_dim = 10
final_hidden_dim = 70
model = BAMGScorer(graph.number_of_nodes('user'), graph.number_of_nodes('item'), user_hidden_dim, item_hidden_dim, final_hidden_dim, NUM_LAYERS)
opt = torch.optim.Adam(model.parameters())
NUM_EPOCHS = 15
for _ in range(NUM_EPOCHS):
model.train()
with tqdm.tqdm(dataloader) as t:
# for pos_pair_graph, neg_pair_graph, blocks in t: # sampler return
for input_nodes, pair_graph, blocks in t:
user_emb, item_emb = model(blocks)
score = model.compute_score(pair_graph, user_emb, item_emb)
loss = compute_margin_loss(score)
opt.zero_grad()
loss.backward()
opt.step()
t.set_postfix({'loss': '%.4f' % loss.item()}, refresh=False)
model.eval()
Is this implementation making use of the embedding learned with nn.Embedding in the process of message passing and aggregation?