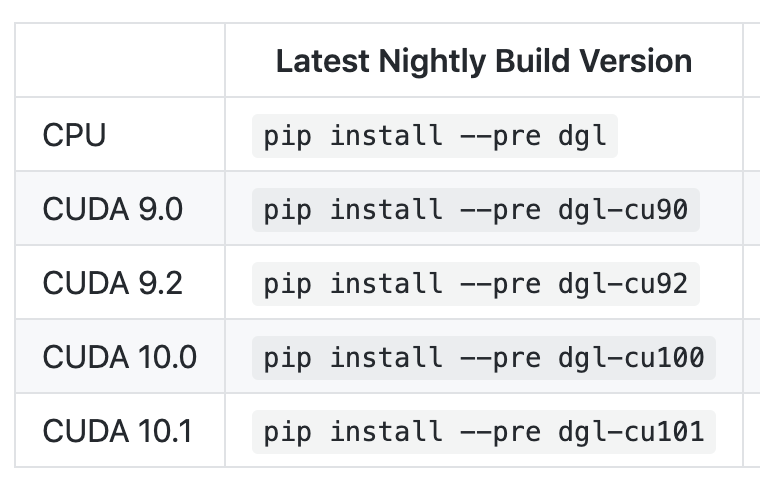
@acho, we have merged DGLGraph and BatchedDGLGraph and provided the flatten api in the master branch. You can try this new feature by install from the source code in the master branch or pip install the nightly build version by:
Now you can add new nodes/edges on the batched graph:
>>> import dgl
>>> import torch
>>> g = dgl.DGLGraph()
>>> g.add_nodes(3)
>>> g.add_edges([0,1,2],[1,2,0])
>>> g.ndata['h'] = torch.ones(3, 5)
>>> g1 = dgl.DGLGraph()
>>> g1.add_nodes(4)
>>> g1.add_edges([0,1,2,3],[0,1,2,3])
>>> g1.ndata['h'] = torch.ones(4, 5) * 2
>>> large_g = dgl.batch([g, g1])
>>> large_g.ndata
{'h': tensor([[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.]])}
>>> large_g.batch_size
2
>>> large_g.batch_num_nodes
[3, 4]
>>> large_g.batch_num_edges
[3, 4]
Note that you can add/remove nodes/edges on large_g directly, but you will receive a warning
>>> large_g.add_nodes(5)
/Users/###/dgl/python/dgl/base.py:25: UserWarning: The graph has batch_size > 1, and mutation would break batching related properties, call `flatten` to remove batching information of the graph.
warnings.warn(msg, warn_type)
To depress the warning, you can call large_g.flatten to make it a single graph:
>>> large_g.flatten()
>>> large_g.batch_size
1
>>> large_g.batch_num_nodes
[7]
>>> large_g.batch_num_edges
[7]
>>> large_g.add_nodes(5)
>>> large_g.ndata
{'h': tensor([[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.],
[2., 2., 2., 2., 2.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.]])}
I hope this could satisfy your needs.