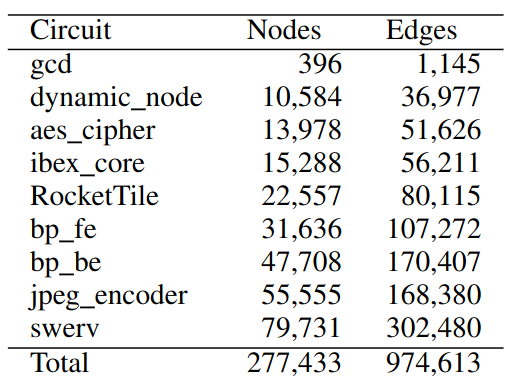
Hi everyone. I have a dataset like following image, with 9 graphs, 277K nodes and 974K edges in total:
I could have other two graphs, although they are too large, and currently I am just removing them.
So far I was performing my experiments with a GATconv from an example I found on the DGL documentation:
class GAT( nn.Module ):
def __init__( self, in_size, hid_size, out_size, heads ):
super().__init__()
self.gat_layers = nn.ModuleList()
self.gat_layers.append( dglnn.GATConv( in_size, hid_size, heads[0], activation = F.relu, allow_zero_in_degree = not SELF_LOOP ))
self.gat_layers.append( dglnn.GATConv( hid_size*heads[0], hid_size, heads[1], residual=True, activation = F.relu, allow_zero_in_degree = not SELF_LOOP ))
self.gat_layers.append( dglnn.GATConv( hid_size*heads[1], out_size, heads[2], residual=True, activation=None, allow_zero_in_degree = not SELF_LOOP ) )
def forward( self, g, inputs ):
h = inputs
for i, layer in enumerate( self.gat_layers ):
h = layer( g, h )
if i == 2: # last layer
h = h.mean(1)
else: # other layer(s)
h = h.flatten(1)
return h
Although I am getting errors saying my GPU has no more free memory. For this reason I would like to try stochastic training, would you say it is a good idea?
I am currently using GraphDataLoader on my dataset, and I can’t even use a batch size of 2:
train_dataset = DataSetFromYosys( currentDir, split, mode='train' )
val_dataset = DataSetFromYosys( currentDir, split, mode='valid' )
test_dataset = DataSetFromYosys( currentDir, split, mode='test' )
train_dataloader = GraphDataLoader( train_dataset, batch_size=1 )
val_dataloader = GraphDataLoader( val_dataset, batch_size=1 )
test_dataloader = GraphDataLoader( test_dataset, batch_size=1 )
Where DataSetFromYosys implements a DGLDataset.
That being said I have some questions:
-
Will I have to use “dgl.dataloading.DataLoader” instead of GraphDataLoader to implement a stochastic training? Shouldn’t GraphDataLoader have a sampler option parameter, like dgl.dataloading.DataLoader does?
-
I got confused with the terminology. What are the batches from GraphDataLoader? when I try to print the nodes of a batch it seems to be a union of graphs from the conventional DGLDataset. Can I make them a mini-batch? Maybe a graph batch and mini-batches are completely different things?
-
What is the difference between a batch and a sampler? I am having a hard time understanding what the sampler is. It seems so similar to a node embedding.
-
I am also trying to change from GATconv to SAGEconv, I believe I read somewhere that SAGE should be lighter when considering memory, is it correct? Why so, where can I understand this better? Modifying the node embedding should help with memory issues, right?
I couldn’t understand what the “train_nids” in this tutorial are about. How can I have a tensor of node IDs if I have multiple graphs? There is no explanation on how to define train_nids, or what it is.