Could you kindly recommend an unsupervised GNN method that could generate edge weight (to rank the importance of a node’ neighbors to the node)?
I assume this is resolved via alternative communication channels.
The answer is that there does not exist research that does exactly what you described. One possibility is to extend previous work on learning node embeddings with unsupervised learning.
Feel free for a follow-up discussion.
SuperGAT
Paper link: How to Find Your Friendly Neighborhood: Graph Attention Design with Self-Supervision
The author designed a self-supervised task (link prediction) to perform unsupervised learning. The e_ij learned by graph attention just means neighbour’s importance.
Thank you for your suggestion! Your implement pytorch-tutorial/train.py at f1400e926e446f348668af7a4449f78c8a50e6f5 · ZZy979/pytorch-tutorial · GitHub is inspiring. However, we do not have label for training, and loss function must change: F.cross_entropy(logits[train_idx], labels[train_idx]). Could you kindly suggest how to change the loss function?
By using attn_loss returned by the model only, the model is trained by link prediction task only, without node labels.
The tensor e here is the edge feature you want. Each item e_ij means the importance of neighbour j to node i for edge <i, j>.
Thank you for your timely reply! I have adapted your code as follows for unsupervised learning.
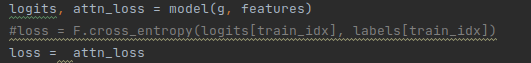
This code has successfully run, however, the loss is around 0.6 in various datasets, such as cora . I have known that unsupervised learning is well-known to be lower accuracy. Might some trick greatly improve the performance?
The attn_loss is from link prediction, while the accuracy is computed for node classification. As you’ve removed classification loss (cross entropy), the model is only trained by link prediction task and can’t learn any information about label.
As the author pointed out in the paper:
… it is hard to learn the relational importance from edges by simply optimizing graph attention for link prediction.
(The “relational importance” here means label information.)
So what do you want to do by unsupervised learning? For supervised downstream task?
- Our data does not contain enough label, therefore, we want to train an unsupervised model. Then we can extract edge weight to select most essential neighbor for each node. Our aim is to select the most essential neighbor, what is your suggestion?
- How to match the edge weight and edge name? In cora dataset, the shape of e is (9228,8,1), while g.edata has no attribute. How do I know which pair of nodes make up an edge?
- Even though you only have a few labled nodes, you can still perform node classification, and it’s helpful for the model to learn better graph attention (neighbour importance).
# suppose -1 means "no label"
train_mask = labels != -1
...
logits, attn_loss = model(g, features)
loss = F.cross_entropy(logits[train_mask], labels[train_mask])
loss += args.attn_loss_weight * attn_loss
As the author puts, node classification guides graph attention to give higher weight to neighbours with the same label, while link prediction helps learn graph structural information.
- The shape of tensor
eis(E, K, 1), and it’s matched with edges by edge id:g.edata['e'] = e.
Actually, it was originally an edge feature:
“K” means the number of attention heads. You can take the mean of each head’s attention value as this:
- In DGL,
g.edges()returns edges ofgas a pair(source nodes, destination nodes), with index corresponding to edge ids.
>>> g = dgl.graph(([0, 0, 1], [1, 2, 2]))
>>> g.edges()
(tensor([0, 0, 1]), tensor([1, 2, 2]))
>>> g.find_edges(eid=0)
(tensor([0]), tensor([1]))
In practice, however, edge features are used in message passing, and you don’t need to know the source and destination nodes of each edge.
Hope my answers make sense to you.
Thank you for your detailed response.
- In fact, our graph composed of two kind of nodes, we have the label of all nodes, our aim is to select the essential neighbor for each node. Also, we have 2 types of edges (relationship), if possible, we want to select the essential neighbor of a node in each type of edges.
- The output in your last suggestion is a tensor composed of numbers. I guess the numbers were ordered according to the input feature, then I just need to match the feature name to the number. Is that the right thing to do?
These numbers are node ids, not input features. What do you mean by “feature name”?
nodetable
featurename type Attr
A type2 5
B type1 3
C type1 2
D type2 5
This is my input node table. Did node ids (0,1,2,3) in tensor of g.edges() is A,B,C,D?
Sorry, but I don’t understand what this table means.
“Node id” has nothing to do with “node feature”.
Thank for your good question on node id, I have managed to figure out it by reading documents and trial and error. One more question, what is the publication source of the unsupervised attr_loss in your implementation of supergat? Is it in the original paper?
Yes, attn_loss is from Eq. (5) in the paper.
This topic was automatically closed 30 days after the last reply. New replies are no longer allowed.
