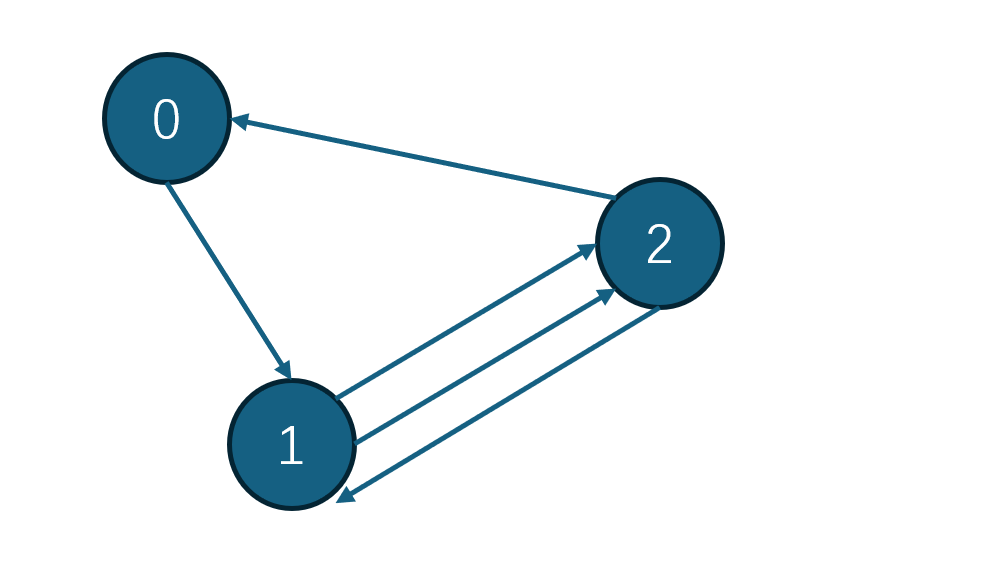
I have a directed graph which also have multiple edges between same node, now I want to use metis to partition the graph.
According to metis manual, metis can only generate partitions for undirected graph and it can’t deal with multiple edges. My solution is first merging multiple edges into one edge with edge weight equal to num of edges. And I will make the directed graph to undirected graph by addding symmetric edges and adding edge weights of edge(u,v) and edge(v, u).
Consider the following example:
What I expect is after the process, there will be one edge between node 1 and node 2 with edge weight equal to 3. I first use dgl.to_simple to remove multiple edge, the use dgl.to_didirected to add symmetrics edges? But how can I add the edge weight of edge(1 → 2) and edge (2 → 1). I’ve checked the doc but didn’t find such function. Do I just have to go through all the edges and add them up.
I will appreciate if someone could give me some advice!
g = dgl.graph(('csr',(tcsr.ind, tcsr.nbr, tcsr.eid)))
g, edge_map = dgl.to_simple(g,return_counts="weight", writeback_mapping=True)
g = dgl.to_bidirected(g)