thanks a lot!
I build a large hetero graph for link prediction, contains two node types : author and research interests; four relation types (author coauthor author) ( author coauthor_rev author)–reverse type since dgl is direct. (author hasinterest interest) (interest researchedby author).
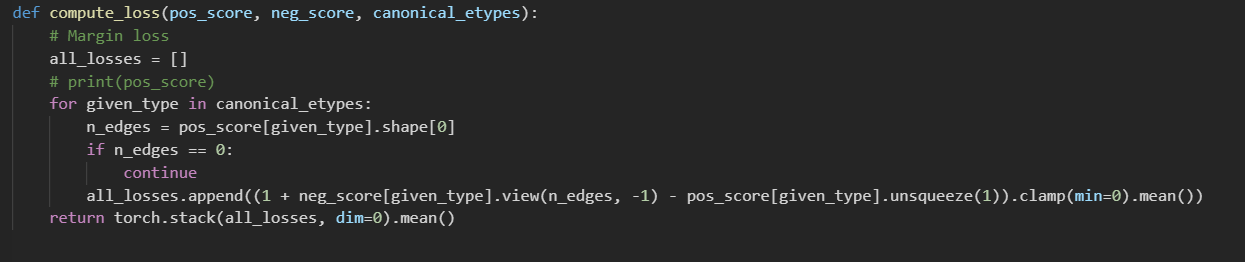
I conduct link prediction training only on edge type (author coauthor author) , I split the (author coauthor author)edge ids in training graph to training set and validation set. should I remove validation edges and their reverse edges from the training graph?
while doing validation, should I use the original graph( which contains all training and validation edges ) for g_sampling or the graph after I remove validation edges and their reverse edges ?
thanks in advance!
I constructed another test graph, which cantains coauthorship in the next year of the training graph,for example training graph is constructed for coauthorship in year 2019, and test graph cantains coauthorship in 2020, train graph and test graph contain exact same set of authors.
and the training graph is splited into train and valid graph. first I chose 10% coauthor edge, use edge_subgraph to extract the subgraph for validation, then I remove the valid edge ids, so valid edge ids are invisible during training.
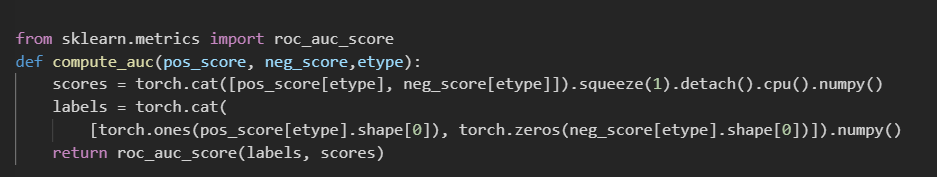
my problem is both test edges and valid edges are invisible during training, but the AUC value on valid edges is 0.9646,and 0.6468 on test edges.