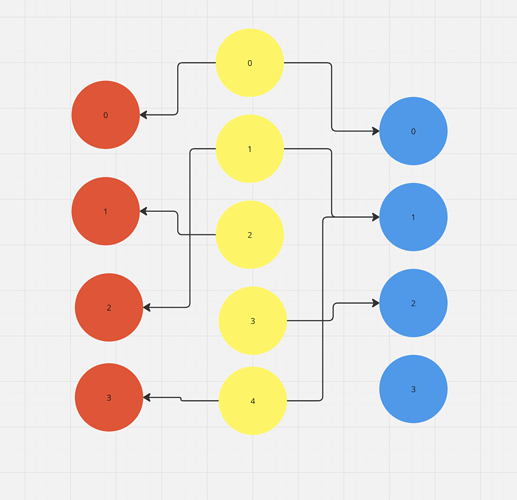
I have a follow up question. I created the following scenario to understand how ondiskdataset loading works:
Imagine the yellow nodes are person nodes, the blue nodes are item nodes and the red nodes are food nodes in the image below:
I recreate this and save as shown in the code below:
import os
import dgl
import torch
import numpy as np
import pandas as pd
# Define the graph structure
# Define directories
base_dir = './sample_dataset'
os.makedirs(base_dir, exist_ok=True)
personNodes = [0, 1, 2, 3, 4]
personNodesWithLinks = [0, 1, 3, 4]
personNodesNp = np.array(personNodesWithLinks)
itemNodes = [0, 1, 2, 3]
foodNodes = [0, 1, 2, 3]
personItemEdges = [[0, 0], [1, 1], [3, 2], [4, 1]]
personItemTotalEdges = [[[0, 0], [0,1]], [[1, 1], [1,2]], [[3, 2], [3, 1]], [[4, 1], [4,3]]]
personItemEdgesNp = np.array(personItemEdges)
personItemTotalEdgesNp = np.array(personItemTotalEdges)
# df = pd.DataFrame(personItemEdgesNp)
# df.to_csv(os.path.join(base_dir, "person_item_edges.csv"), index=False, header=False)
np.save(os.path.join(base_dir, "person_item_edges.npy"), personItemEdgesNp)
np.save(os.path.join(base_dir, "person_item_edges_t.npy"), personItemEdgesNp.T)
np.save(os.path.join(base_dir, "person_nodes.npy"), personNodesNp)
np.save(os.path.join(base_dir, "person_item_edges_total.npy"), personItemTotalEdgesNp)
personFoodEdges = [[0, 0], [1, 2], [2, 1], [4, 3]]
personFoodEdgesNp = np.array(personFoodEdges)
# df = pd.DataFrame(personFoodEdgesNp)
# df.to_csv(os.path.join(base_dir, "person_food_edges.csv"), index=False, header=False)
np.save(os.path.join(base_dir, "person_food_edges.npy"), personFoodEdgesNp)
np.save(os.path.join(base_dir, "person_food_edges_t.npy"), personFoodEdgesNp.T)
# Add features to nodes (dummy features for this example)
features = torch.eye(len(personNodes)) # Identity matrix as features
# Save node features
torch.save(features, os.path.join(base_dir, "node_features.pt"))
# Create the metadata.yaml file
yaml_content = f"""
dataset_name: sample_graph
graph:
nodes:
- type: person
num: {len(personNodes)}
- type: item
num: {len(itemNodes)}
- type: food
num: {len(foodNodes)}
edges:
- type: "person:like:item"
format: numpy
path: ./person_item_edges_t.npy
- type: "person:like:food"
format: numpy
path: ./person_food_edges_t.npy
feature_data:
- domain: node
type: person
name: feature
format: torch
in_memory: false
path: node_features.pt
tasks:
- name: link_prediction
train_set:
- type: "person:like:item"
data:
- name: seeds
format: numpy
in_memory: false
path: ./person_item_edges.npy
- name: seeds_with_negative
format: numpy
in_memory: false
path: ./person_item_edges_total.npy
"""
# Save metadata.yaml
with open(os.path.join(base_dir, "metadata.yaml"), "w") as f:
f.write(yaml_content)
I have saved negative pairs as seeds_with_negative. Now I am loading the dataset as in the documentation for link prediction but removing the negative sampling as suggested. The code is shown below:
base_dir = './sample_dataset'
print(base_dir)
dataset = gb.OnDiskDataset(base_dir).load()
itemset = dataset.tasks[0].train_set
graph = dataset.graph
print(itemset)
print(graph)
sampler = gb.ItemSampler(
itemset,
batch_size=1,
# minibatcher=minibatcher,
shuffle=True,
)
sampler = sampler.sample_neighbor(
graph, [-1]
)
for minibatch in sampler:
print(minibatch)
I am seeing the following output when I print the minibatch after sample_neighbours. As you can see, the person:like:food edges are showing as 0 everywhere. Why is this the case? Given that I use a fanout of -1, I should be seeing some edges in at least one of the minibatches for this edge type.
MiniBatch(seeds_with_negative={'person:like:item': tensor([[[4, 1],
[4, 3]]])},
seeds={'person:like:item': tensor([[4, 1]], dtype=torch.int32)},
sampled_subgraphs=[SampledSubgraphImpl(sampled_csc={'person:like:food': CSCFormatBase(indptr=tensor([0], dtype=torch.int32),
indices=tensor([], dtype=torch.int32),
), 'person:like:item': CSCFormatBase(indptr=tensor([0, 2], dtype=torch.int32),
indices=tensor([1, 0], dtype=torch.int32),
)},
original_row_node_ids={'person': tensor([4, 1], dtype=torch.int32)},
original_edge_ids=None,
original_column_node_ids={'food': tensor([], dtype=torch.int32), 'item': tensor([1], dtype=torch.int32), 'person': tensor([4], dtype=torch.int32)},
)],
node_features=None,
labels=None,
input_nodes={'person': tensor([4, 1], dtype=torch.int32)},
indexes=None,
edge_features=None,
compacted_seeds={'person:like:item': tensor([[0, 0]], dtype=torch.int32)},
blocks=[Block(num_src_nodes={'food': 0, 'item': 0, 'person': 2},
num_dst_nodes={'food': 0, 'item': 1, 'person': 1},
num_edges={('person', 'like', 'food'): 0, ('person', 'like', 'item'): 2},
metagraph=[('person', 'food', 'like'), ('person', 'item', 'like')])],
)
MiniBatch(seeds_with_negative={'person:like:item': tensor([[[1, 1],
[1, 2]]])},
seeds={'person:like:item': tensor([[1, 1]], dtype=torch.int32)},
sampled_subgraphs=[SampledSubgraphImpl(sampled_csc={'person:like:food': CSCFormatBase(indptr=tensor([0], dtype=torch.int32),
indices=tensor([], dtype=torch.int32),
), 'person:like:item': CSCFormatBase(indptr=tensor([0, 2], dtype=torch.int32),
indices=tensor([0, 1], dtype=torch.int32),
)},
original_row_node_ids={'person': tensor([1, 4], dtype=torch.int32)},
original_edge_ids=None,
original_column_node_ids={'food': tensor([], dtype=torch.int32), 'item': tensor([1], dtype=torch.int32), 'person': tensor([1], dtype=torch.int32)},
)],
node_features=None,
labels=None,
input_nodes={'person': tensor([1, 4], dtype=torch.int32)},
indexes=None,
edge_features=None,
compacted_seeds={'person:like:item': tensor([[0, 0]], dtype=torch.int32)},
blocks=[Block(num_src_nodes={'food': 0, 'item': 0, 'person': 2},
num_dst_nodes={'food': 0, 'item': 1, 'person': 1},
num_edges={('person', 'like', 'food'): 0, ('person', 'like', 'item'): 2},
metagraph=[('person', 'food', 'like'), ('person', 'item', 'like')])],
)
MiniBatch(seeds_with_negative={'person:like:item': tensor([[[3, 2],
[3, 1]]])},
seeds={'person:like:item': tensor([[3, 2]], dtype=torch.int32)},
sampled_subgraphs=[SampledSubgraphImpl(sampled_csc={'person:like:food': CSCFormatBase(indptr=tensor([0], dtype=torch.int32),
indices=tensor([], dtype=torch.int32),
), 'person:like:item': CSCFormatBase(indptr=tensor([0, 1], dtype=torch.int32),
indices=tensor([0], dtype=torch.int32),
)},
original_row_node_ids={'person': tensor([3], dtype=torch.int32)},
original_edge_ids=None,
original_column_node_ids={'food': tensor([], dtype=torch.int32), 'item': tensor([2], dtype=torch.int32), 'person': tensor([3], dtype=torch.int32)},
)],
node_features=None,
labels=None,
input_nodes={'person': tensor([3], dtype=torch.int32)},
indexes=None,
edge_features=None,
compacted_seeds={'person:like:item': tensor([[0, 0]], dtype=torch.int32)},
blocks=[Block(num_src_nodes={'food': 0, 'item': 0, 'person': 1},
num_dst_nodes={'food': 0, 'item': 1, 'person': 1},
num_edges={('person', 'like', 'food'): 0, ('person', 'like', 'item'): 1},
metagraph=[('person', 'food', 'like'), ('person', 'item', 'like')])],
)
MiniBatch(seeds_with_negative={'person:like:item': tensor([[[0, 0],
[0, 1]]])},
seeds={'person:like:item': tensor([[0, 0]], dtype=torch.int32)},
sampled_subgraphs=[SampledSubgraphImpl(sampled_csc={'person:like:food': CSCFormatBase(indptr=tensor([0], dtype=torch.int32),
indices=tensor([], dtype=torch.int32),
), 'person:like:item': CSCFormatBase(indptr=tensor([0, 1], dtype=torch.int32),
indices=tensor([0], dtype=torch.int32),
)},
original_row_node_ids={'person': tensor([0], dtype=torch.int32)},
original_edge_ids=None,
original_column_node_ids={'food': tensor([], dtype=torch.int32), 'item': tensor([0], dtype=torch.int32), 'person': tensor([0], dtype=torch.int32)},
)],
node_features=None,
labels=None,
input_nodes={'person': tensor([0], dtype=torch.int32)},
indexes=None,
edge_features=None,
compacted_seeds={'person:like:item': tensor([[0, 0]], dtype=torch.int32)},
blocks=[Block(num_src_nodes={'food': 0, 'item': 0, 'person': 1},
num_dst_nodes={'food': 0, 'item': 1, 'person': 1},
num_edges={('person', 'like', 'food'): 0, ('person', 'like', 'item'): 1},
metagraph=[('person', 'food', 'like'), ('person', 'item', 'like')])],
)