I have more than 100 full connected networks, and their edges were undirectional and weighted, how to quantify and classify those networks based on their node and edge attributes? Each network represents one patient, we hope to unsupervised classify those networks.
This question was on graph Isomorphism or graph classification to binary classify the health and disease patients. Our main focus is mainly on edge attributes. Any related survey or algorithm integrating edge attribute would be helpful! Thank you for your help.
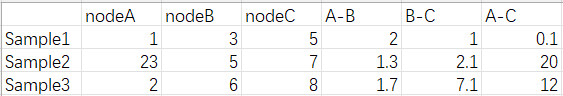
An example, our graphs have numeric node and edge attributes, no label.