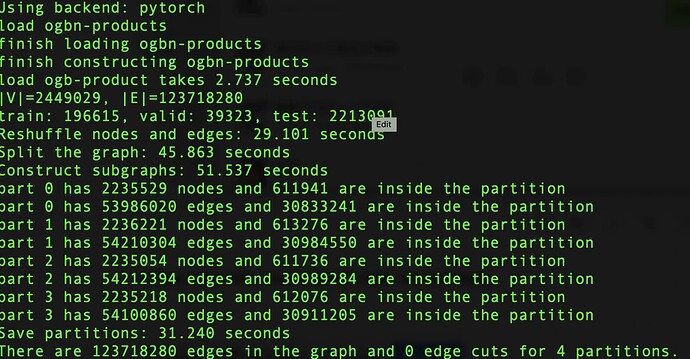
Great library! I am currently learning distributed training of GNN with DGL. I have tested both metis and random partition methods and surprisingly found out that the random partition method always returns a graph partition with 0 edge cut (ogbn-proteins/ogbn-products dataset with 10000 partitions). It seems the random partition only assigns each node to different partitions with uniform probability, so I am confused as to why the edge cut is still 0.
Any help would be appreciated!