Hi everyone! I’m stuck on my project for a month.
More precisely, I’m trying to train my GAT model over a single graph for node classification.
Nodes have a feature vector of 20 elements, and a label (0,1,2,3).
I saved my DGL graph in a bin with all the information. When I need to train, I load the graph using dgl.
The problem is that I expect that since Im using a single Graph (instead than all the graphs) the Model should overfit because I’m using only it as a sample. This doesn’t happen.
My graph contains 2k nodes and the relation is ‘Adjacency’ so if A is adjacent to B then they will be related to each other.
This is my model:
import torch
import torch.nn as nn
import torch.optim as optim
import dgl.nn as dglnn
import torch.nn.functional as F
class GATLayer(nn.Module):
def __init__(self, in_features, out_features, num_heads):
super(GATLayer, self).__init__()
self.gat = dglnn.GATConv(in_feats=in_features, out_feats=out_features, num_heads=num_heads, allow_zero_in_degree=True)
def forward(self, graph, inputs):
return self.gat(graph, inputs)
class GATDummy(nn.Module):
def __init__(self, in_features, hidden_features, num_heads, num_classes):
super(GATDummy, self).__init__()
self.layer1 = GATLayer(in_features, hidden_features, num_heads)
self.layer2 = GATLayer(hidden_features * num_heads, hidden_features, 2)
self.layer3 = GATLayer(hidden_features * 2, hidden_features, 2)
self.layer4 = GATLayer(hidden_features * 2, hidden_features, 1)
self.layer5 = GATLayer(hidden_features, num_classes, 1)
def forward(self, graph, inputs):
h = self.layer1(graph, inputs)
h = F.elu(h)
h = h.view(h.shape[0], -1)
h = self.layer2(graph, h)
h = F.elu(h)
h = h.view(h.shape[0], -1)
h = self.layer3(graph, h)
h = F.elu(h)
h = h.view(h.shape[0], -1)
h = self.layer4(graph, h)
h = F.elu(h)
h = h.view(h.shape[0], -1)
h = self.layer5(graph, h)
return h.view(h.shape[0], -1)
I’m using 256 as hidden features.
My training function is this:
def train(dgl_train_graphs, dgl_validation_graphs, model, loss_w):
# Define the optimizer
optimizer = optim.AdamW(model.parameters(), lr=0.0001)
print('Training started...')
for e in range(500):
model.train()
total_loss = 0
total_recall = 0
total_precision = 0
total_train_f1 = 0
for g in dgl_train_graphs:
# Get the features, labels, and masks for the current graph
features = g.ndata["feat"]
labels = g.ndata["label"]
# 1,2,3,4 -> 0,1,2,3
labels = labels - 1
# Forward pass
logits = model(g, features)
# Compute prediction
pred = torch.argmax(logits, dim=1)
# Compute loss with class weights
criterion = torch.nn.CrossEntropyLoss(weight=loss_w)
loss = criterion(logits, labels)
# 0,1,2,3 -> 1,2,3,4
pred = pred + 1
labels = labels + 1
recall, precision, f1 = compute_metrics(pred, labels)
# Accumulate loss and accuracy values for this graph
total_loss += loss.item()
total_recall += recall.item()
total_precision += precision.item()
total_train_f1 += f1.item()
# Backward pass
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(f"EPOCH {e} | loss: {avg_loss:.3f} | precision train WT: {avg_train_precision:.3f} | f1-score train WT: {avg_train_f1:.3f}|")
This are my results:
EPOCH 6 | loss: 2.557 | precision train WT: 0.041 | f1-score train WT: 0.079|
EPOCH 7 | loss: 2.736 | precision train WT: 0.041 | f1-score train WT: 0.079|
EPOCH 8 | loss: 1.935 | precision train WT: 0.041 | f1-score train WT: 0.079|
EPOCH 9 | loss: 1.500 | precision train WT: 0.044 | f1-score train WT: 0.085|
...
EPOCH 132 | loss: 1.158 | precision train WT: 0.091 | f1-score train WT: 0.166|
EPOCH 133 | loss: 1.166 | precision train WT: 0.098 | f1-score train WT: 0.173|
EPOCH 134 | loss: 1.183 | precision train WT: 0.083 | f1-score train WT: 0.153|
EPOCH 135 | loss: 1.182 | precision train WT: 0.099 | f1-score train WT: 0.173|
EPOCH 136 | loss: 1.162 | precision train WT: 0.086 | f1-score train WT: 0.158|
...
EPOCH 493 | loss: 0.637 | precision train WT: 0.105 | f1-score train WT: 0.189|
EPOCH 494 | loss: 0.639 | precision train WT: 0.126 | f1-score train WT: 0.223|
EPOCH 495 | loss: 0.635 | precision train WT: 0.104 | f1-score train WT: 0.187|
EPOCH 496 | loss: 0.629 | precision train WT: 0.118 | f1-score train WT: 0.210|
EPOCH 497 | loss: 0.629 | precision train WT: 0.118 | f1-score train WT: 0.210|
EPOCH 498 | loss: 0.631 | precision train WT: 0.105 | f1-score train WT: 0.190|
EPOCH 499 | loss: 0.628 | precision train WT: 0.120 | f1-score train WT: 0.213|
Im very frustrated and I can’t understand what is wrong. I checked my features and they are meaningful, look this picture:
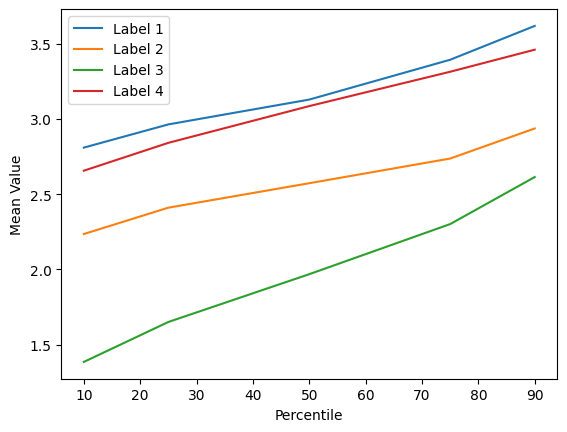
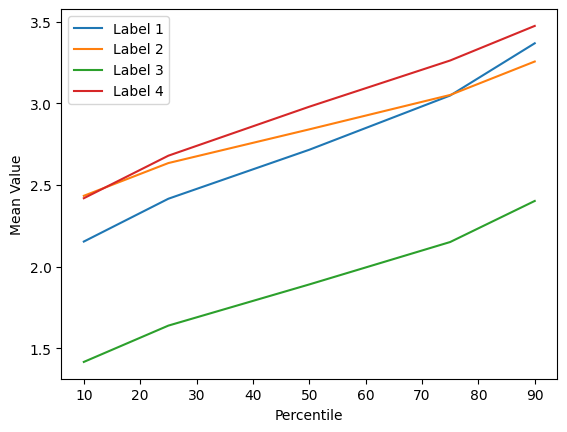
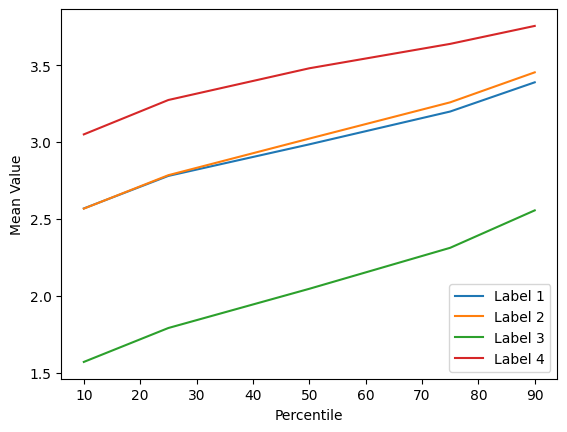
Label: 1,2,4 are tumoral nodes
Label: 3 are non tumoral nodes
Why my model is so weak?
