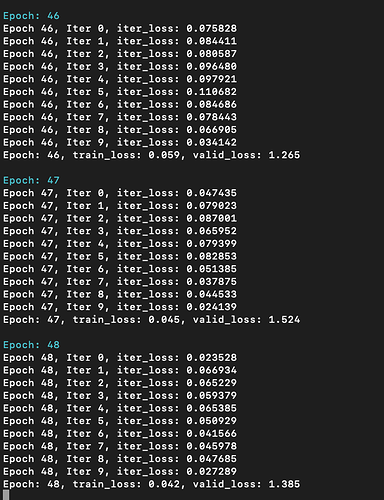
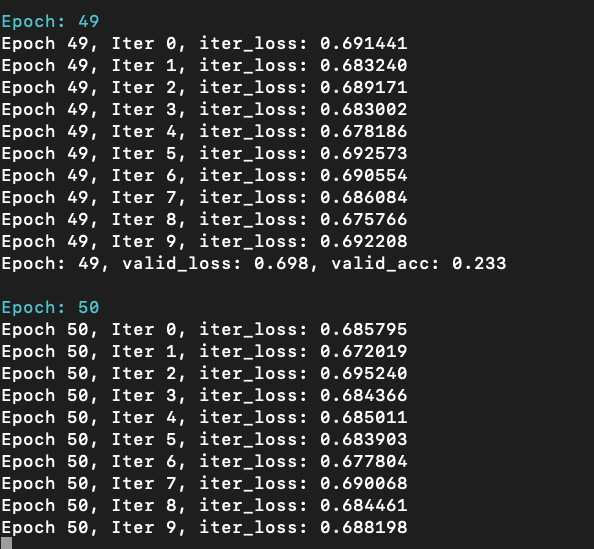
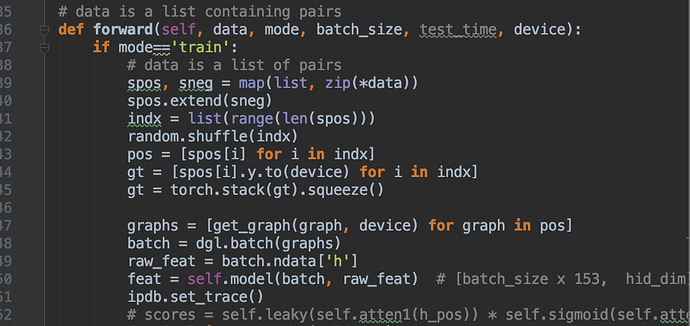
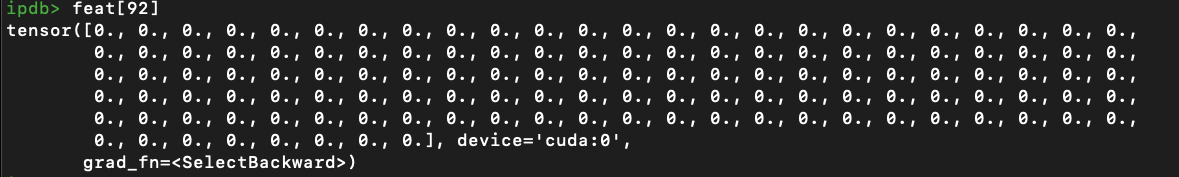
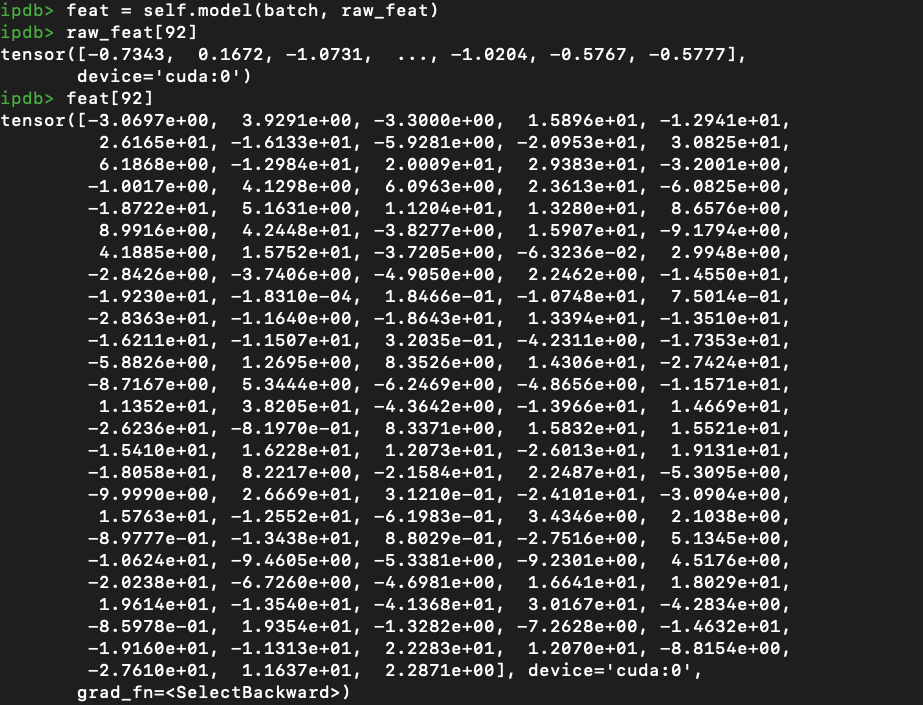
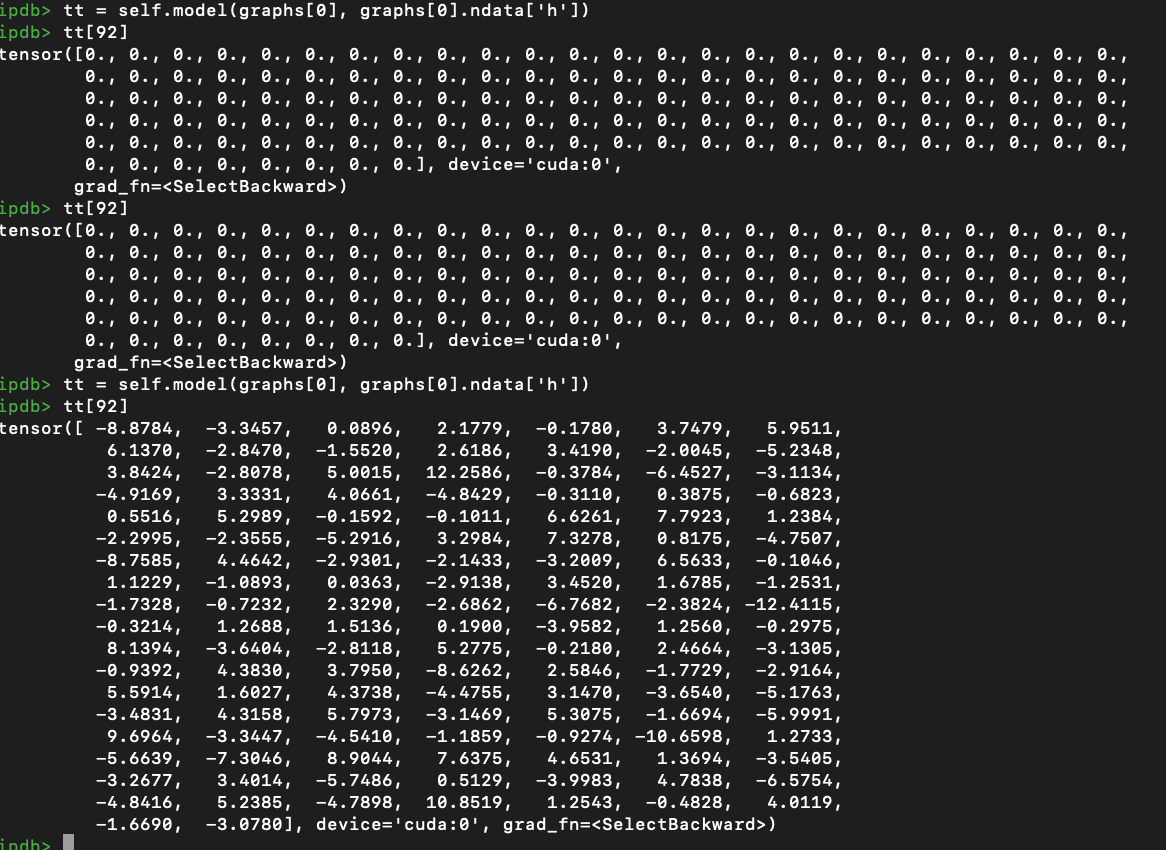
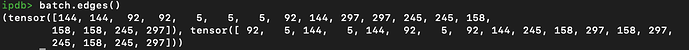I was training on a self-collected dataset using GCN model.
Specifically, I constructed many small graphs batched together during training, with batch_size=100.
For each node, I extracted Resnet 2048-dim feature, the goal is to do binary classification for each graph.
Loss: BCELoss
GCN model:
Connection: bidirectional, fully connected edges for each graph
Model: 3 layers: 2048->128->1, all are GCN layer
MLP model:
For verification, I trained MLP (2048->128->128->1, ReLU in between) on this dataset, it quickly overfits:
whereas GCN model cannot:
Any suggestions? Thanks in advance.