Hi dgl community,
I have a question on how graphSAGE works.
I am currently using it with the SAGEConv module from dgl library.
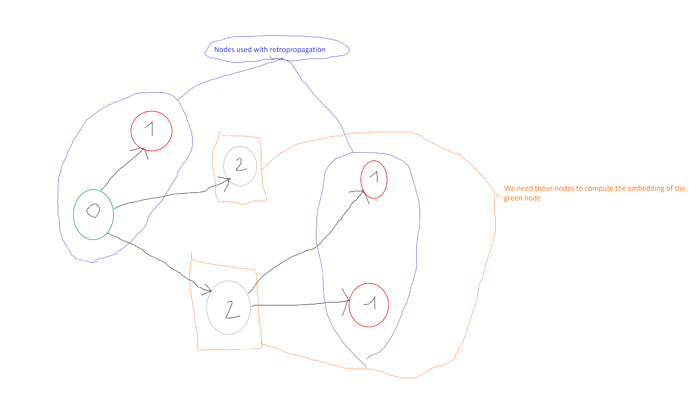
I have a graph of nodes with 3 different labels (0,1 and 2). From this three different labels, I only use two of them to calculate the loss, let say those labels are 0 and 1. Hence, I have a binary classification problem. The thing is, I need the embeddings of the third unused (label : 2) label to generates embeddings of the nodes which have one of the two others labels 0 or 1.
So, in dgl.dataloading.DataLoader I only put the nodes with labels 0 and 1. But to compute the output embedding of a node v (with labels 1 or 0) at iteration K = 2, for instance, I need the embedding of some node u (with label 0, 1 or 2). What I don’t know is how SAGEConv computes the embeddings of the nodes with labels 2 at the iteration K > 1, knowing that they are not included in the dataloader, to cumpute the embedding of node v ?
To summarize, I only train my SAGEConv model on the labels 0 and 1, but I need the label 2 to compute embeddings of the labels 0 and 1.
I hope my question is clear.
Thank you all !