Hi,
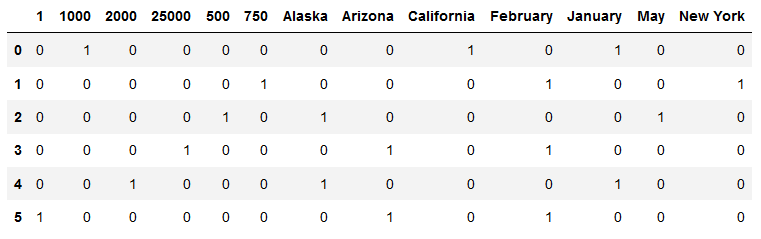
I’m looking for an efficient way to construct a graph from a dataframe of multi-label binarized features. The objective is to obtain a graph in which all the nodes that share the same features are connected. For example, in the df below, there would be edges betweeen the nodes 1, 3 and 5, because they all have the feature ‘February’.
I was thinking of something like iterating through feature names, creating permutation grids of all the nodes that mention them and then concatenating the grids into the final arrays of source and destination nodes. Is there be a better way to do it?