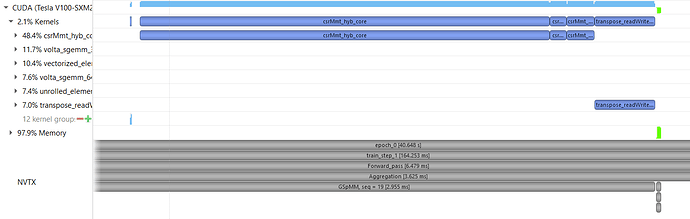
I am running a graphsage model on a single gpu, I understand that CSRMMT kernel can be seen during aggregation step with the update_all() function. But can someone explain why do I see 3 CSRMMT kernels for each layer with GSPMM op. Also what is the reason of transpose_readwrite_alignment kernel being called?
Hi, I got exactly the same question as yours. I wonder if you have figured it out or anyone has an idea about it.
The point is, if there are two sage conv layers, it is supposed to have 2 SpMM calls in each forward pass. But the nvprof shows that there are three calls to the “csrMmt_hyb_core” kernel.
For the transpose kernel, this is because the cusparseScsrmm2 API in CUDA 10 assumes the tensor is column-major and DGL’s feature tensor is row-major, we need to transpose the output matrix. If you use CUDA 11, then you will not encounter this problem because the new cusparseSpMM API with ALG2 assumes the tensor is row-major.
@chenxuhao for three csrMmt_hyb_core calls, this is because a single cusparseScsrmm2 call will trigger several csrMmt_hyb_core kernels. I suggest you upgrading your cuda to version 11 where the new API will only trigger a single csrmm_v2_kernel, and CUDA 11’s spmm is faster.
btw, I noticed that you were editing the binary_reduce_sum.cu in your fork, however, this is legacy code that has been deprecated since dgl 0.5, please edit this file:
Hi Zihao,
Thank you so much for your reply.
I tried CUDA11.1 and it is faster than CUDA10.2 for SAGE.
But I am still confused about the fact that SAGE (mean aggregator) is much slower than GCN.
For example, when running GCN, I see 4 CusparseCsrmm2 calls per epoch (2 in FW and 2 in BW), and one epoch takes 90ms. When I run SAGE-mean, however, I see only 3 CusparseCsrmm2 calls per epoch (2 in FW and 1 in BW), but one epoch takes 400ms.
Note that the epoch time difference is mostly due to the CusparseCsrmm2 calls (i.e. cusparse::csrmm_v2_kernel).
Computation wise, GCN and SAGE-mean only differ in the normalization scores (i.e. Cij for each edge[i,j]). If normalization scores are pre-computed, the CusparseCsrmm2 calls should be exactly the same and thus the speed should be similar for GCN and SAGE-mean, right?
Also, this speed difference between GCN and SAGE is not only on GPU. I see this on CPU as well.
Seems like the spmm kernel calls for GCN and SAGE are different in some sense. But I checked that the parameters for the calls are the same: e.g. for reddit graph, m = 232965 n = 16 k = 232965 nnz = 114615892
Am I missing something?
Best,
Xuhao
Please notice that there was a optimization in our GCNConv implementation:
What it did basically is to determine whether to compute AXW in ((AX)W) or (A(XW)), in some cases the later way is better and I don’t think we have such optimization for GraphSAGE. Would you mind checking if it’s the reason?
This makes sense to me. What prevents you to do this optimization for SAGE? Is it something fundamental or you just didn’t do it?
I think it’s just because GraphSAGE has multiple kinds of aggregators (mean/pool/lstm/…), however for mean aggregator such optimization is appilcable. I’ll create a PR adding this feature.