Hi,
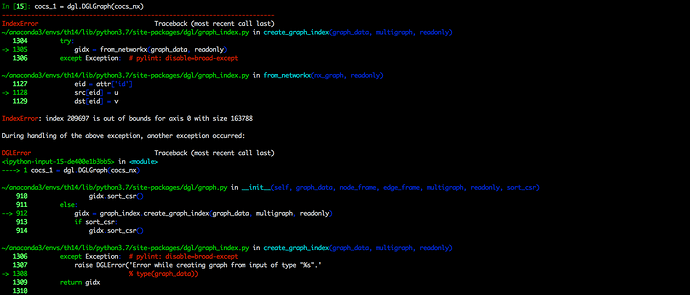
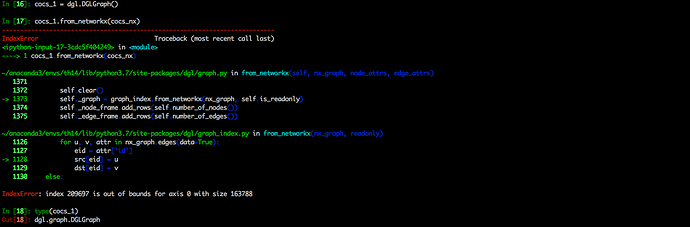
I encountered an error when trying converting a graph of the type <class ‘networkx.classes.digraph.DiGraph’> to a dgl graph via dgl.DGLgraph() function or .from_networkx() method of a dgl.graph.DGLGraph object. The graph is given by calling the .to_networkx() method of another dgl.graph.DGLGraph object.
To be short, the error occured when I try converting networkx graph given by a dgl graph to a new dgl graph.
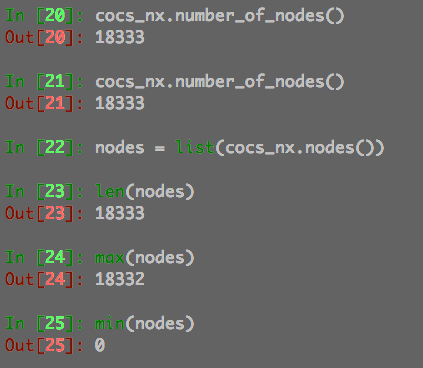
You may see the details from the attached following screenshots.
Hope you can help me! Thanks!