Hello,
Thanks for the quick response.
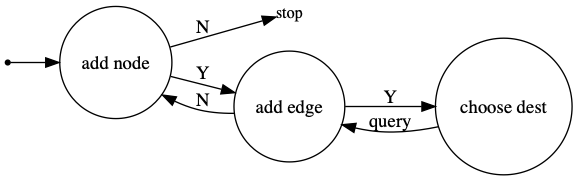
I noticed above code segment. However, I want to understand the general pattern of producing such sequence for a set of temporal edges. You can correct me in the following example:
Edge list have the temporal edges in the chronological order, where first two columns represent the nodes, third column represents the edge type, and the last column is the timestamp.
edge_list = [[0,1,"Type_A",10],
[1,2,"Type_A",20],
[0,2,"Type_B",30]]
add_set=set()
decision_sequence=[]
for edge in edge_list:
id1=edge[0]
id2=edge[1]
edge_type=edge[2]
if id1 not in add_set:
decision_sequence.append((0,node_type_to_id[id1])) ## Add Node
add_set.add(id1)
if id2 not in add_set:
decision_sequence.append((0,node_type_to_id[id2])) ## Add Node
add_set.add(id2)
decision_sequence.append((1,edge_type_to_id[edge_type])) ## Add Edge Type
decision_sequence.append((2,id2)) ## Add destination
decision_sequence.append((1,no_of_edge_types)) ## Stop Adding Edge
decision_sequence.append((0,no_of_node_types)) ## Stop Adding Nodes
node_type_to_id and edge_type_to_id are dictionaries that map given node or edge to an arbitrary type.