Hi, I understand that the DGL graph I am transforming to Networkx using .to_networkx is not a multigraph, although you can see on the print value that it is a multigraph. Is this behavior expected? Here is my code snippet:
drawGraph( self.graph, self.graph.name )
nx_graph = self.graph.to_networkx()
print( "nx_graph:\n", nx_graph )
print( ".nodes:\n", nx_graph.nodes(data=True))
print( ".edges:\n", nx_graph.edges(data=True))
eigen = nx.eigenvector_centrality( nx_graph )
Here is the output I am getting:
nx_graph:
MultiDiGraph with 14 nodes and 15 edges
.nodes:
[(0, {}), (1, {}), (2, {}), (3, {}), (4, {}), (5, {}), (6, {}), (7, {}), (8, {}), (9, {}), (10, {}), (11, {}), (12, {}), (13, {})]
.edges:
[(2, 11, {'id': 0}), (3, 13, {'id': 1}), (4, 8, {'id': 2}), (4, 13, {'id': 3}), (5, 9, {'id': 4}), (5, 11, {'id': 5}), (6, 8, {'id': 6}), (7, 1, {'id': 7}), (8, 7, {'id': 8}), (9, 7, {'id': 9}), (10, 0, {'id': 10}), (11, 10, {'id': 11}), (12, 10, {'id': 12}), (13, 9, {'id': 13}), (13, 12, {'id': 14})]
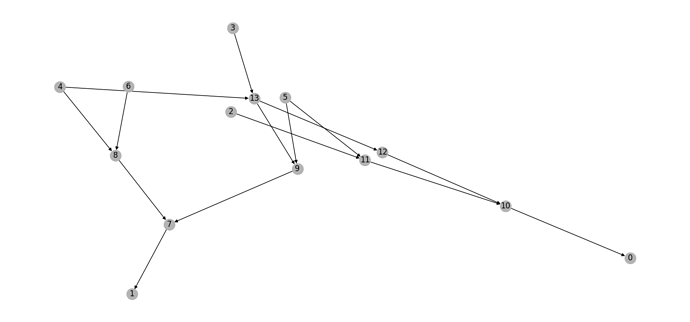
Here is an image of the graph in self.graph from drawGraph():
Furthermore I am getting an error when trying to calculate the eigenvector centrality because it is a multigraph:
Traceback (most recent call last):
File "/home/gudeh/Desktop/congestionPrediction/regression.py", line 1056, in <module>
train_dataset = DataSetFromYosys( currentDir, split, ablationIter, mode='train' )
File "/home/gudeh/Desktop/congestionPrediction/regression.py", line 267, in __init__
super().__init__( name='mydata_from_yosys_'+mode )
File "/home/gudeh/.local/lib/python3.10/site-packages/dgl/data/dgl_dataset.py", line 112, in __init__
self._load()
File "/home/gudeh/.local/lib/python3.10/site-packages/dgl/data/dgl_dataset.py", line 203, in _load
self.process()
File "/home/gudeh/Desktop/congestionPrediction/regression.py", line 271, in process
graph = self._process_single( path )
File "/home/gudeh/Desktop/congestionPrediction/regression.py", line 347, in _process_single
eigen = nx.eigenvector_centrality( nx_graph )
File "/home/gudeh/.local/lib/python3.10/site-packages/networkx/classes/backends.py", line 148, in wrapper
return func(*args, **kwds)
File "/home/gudeh/.local/lib/python3.10/site-packages/networkx/utils/decorators.py", line 766, in func
return argmap._lazy_compile(__wrapper)(*args, **kwargs)
File "<class 'networkx.utils.decorators.argmap'> compilation 4", line 3, in argmap_eigenvector_centrality_1
File "/home/gudeh/.local/lib/python3.10/site-packages/networkx/utils/decorators.py", line 86, in _not_implemented_for
raise nx.NetworkXNotImplemented(errmsg)
networkx.exception.NetworkXNotImplemented: not implemented for multigraph type