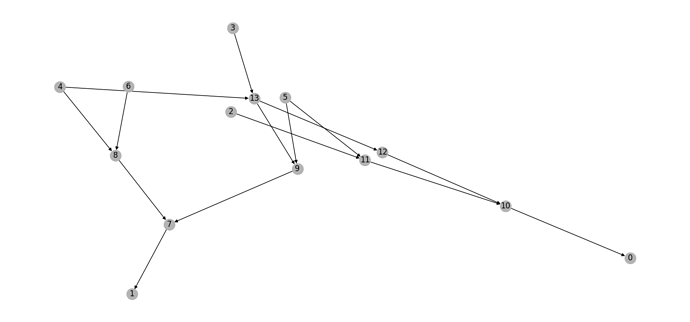
Hi there. I used a piece of code from other post on the forum to remove isolated nodes. You can see a printscreen of a small graph after isolated nodes have been removed. It seems to not have any 0-in-degree nodes as DGL claims.
When I try to perform training with this graph DGL outputs:
File “/home/gudeh/.local/lib/python3.10/site-packages/dgl/nn/pytorch/conv/gatconv.py”, line 254, in forward
raise DGLError('There are 0-in-degree nodes in the graph, ’
dgl._ffi.base.DGLError: There are 0-in-degree nodes in the graph, output for those nodes will be invalid. This is harmful for some applications, causing silent performance regression. Adding self-loop on the input graph by callingg = dgl.add_self_loop(g)will resolve the issue. Settingallow_zero_in_degreeto beTruewhen constructing this module will suppress the check and let the code run.
I am using the same model definition as the PPI example (class GAT), although I have to add “allow_zero_in_degree=True” for the code to work.
From the documentation on GATconv it should be bad for training to have 0-in-degree nodes, so I would like to not have them, if possible. What can I do?
class GAT( nn.Module ):
def __init__( self, in_size, hid_size, out_size, heads ):
super().__init__()
self.gat_layers = nn.ModuleList()
print("\n\nINIT GAT!!")
print("in_size:", in_size)
print("hid_size:", hid_size)
for k,head in enumerate( heads ):
print("head:", k,head)
print("out_size", out_size)
self.gat_layers.append( dglnn.GATConv( in_size, hid_size, heads[0], activation=F.elu, allow_zero_in_degree=True ) )
self.gat_layers.append( dglnn.GATConv( hid_size*heads[0], hid_size, heads[1], residual=True, activation=F.elu, allow_zero_in_degree=True ) )
self.gat_layers.append( dglnn.GATConv( hid_size*heads[1], out_size, heads[2], residual=True, activation=None, allow_zero_in_degree=True ) )
def forward( self, g, inputs ):
h = inputs
for i, layer in enumerate( self.gat_layers ):
h = layer( g, h )
if i == 2: # last layer
h = h.mean(1)
else: # other layer(s)
h = h.flatten(1)
return h