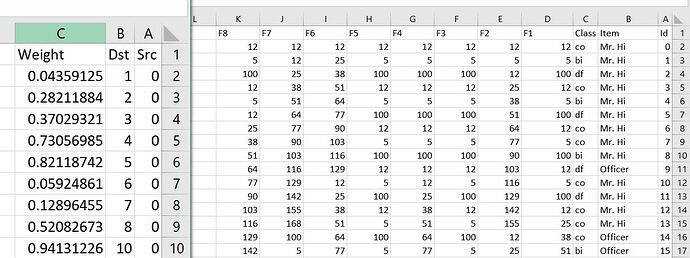
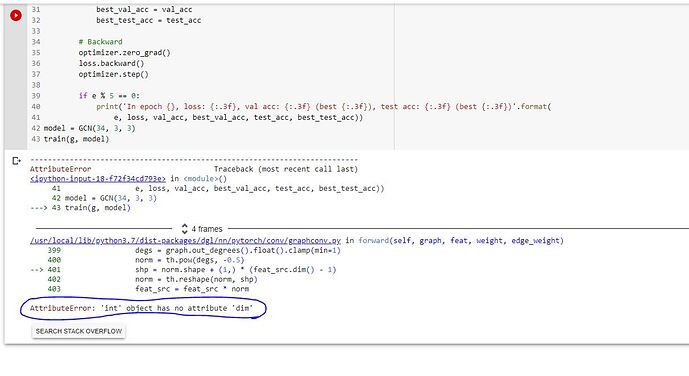
Hi, I got the same issue. Here is what I’m getting. Here is my data loader script.
import dgl
from dgl.data import DGLDataset
import torch
import os
import pandas as pd
from utils import POS2INDEX
import numpy as np
class LabDataset(DGLDataset):
def __init__(self):
super().__init__(name='lab')
def process(self):
nodes_data = pd.read_csv('./members.csv')
edges_data = pd.read_csv('./relations.csv')
node_features = torch.from_numpy(nodes_data['lab'].to_numpy())
node_labels = torch.from_numpy(nodes_data['dpd'].to_numpy())
edge_features = torch.from_numpy(np.array(edges_data['type']))
edges_src = torch.from_numpy(edges_data['src'].to_numpy())
edges_dst = torch.from_numpy(edges_data['dst'].to_numpy())
num_classes = (len(POS2INDEX.values(),))
self.graph = dgl.graph((edges_src, edges_dst), num_nodes=nodes_data.shape[0])
self.graph.ndata['feat'] = node_features
self.graph.ndata['label'] = node_labels
self.graph.edata['edges'] = edge_features
# If your dataset is a node classification dataset, you will need to assign
# masks indicating whether a node belongs to training, validation, and test set.
n_nodes = nodes_data.shape[0]
n_train = int(n_nodes * 0.6)
n_val = int(n_nodes * 0.2)
train_mask = torch.zeros(n_nodes, dtype=torch.bool)
val_mask = torch.zeros(n_nodes, dtype=torch.bool)
test_mask = torch.zeros(n_nodes, dtype=torch.bool)
train_mask[:n_train] = True
val_mask[n_train:n_train + n_val] = True
test_mask[n_train + n_val:] = True
self.graph.ndata['train_mask'] = train_mask
self.graph.ndata['val_mask'] = val_mask
self.graph.ndata['test_mask'] = test_mask
self.num_classes = num_classes
def __getitem__(self, i):
return self.graph
def __len__(self):
return 1
dataset = LabDataset()
graph = dataset[0]
print(graph)
Here is my two-layer GNN
import dgl
import torch
import torch.nn as nn
import torch.nn.functional as F
from dgl.nn import GraphConv
import dgl.data
from load_data import LabDataset
dataset = LabDataset()
print('Number of categories:', dataset.num_classes)
g = dataset[0]
g = g.to('cuda')
g = dgl.add_self_loop(g)
# g = dgl.add_self_loop(g)
print('Node features')
print(g.ndata)
print('Edge features')
print(g.edata)
class GCN(nn.Module):
def __init__(self, in_feats, h_feats, num_classes):
super(GCN, self).__init__()
self.conv1 = GraphConv(in_feats, h_feats)
self.conv2 = GraphConv(h_feats, num_classes)
def forward(self, g, in_feat):
h = self.conv1(g, in_feat)
h = F.relu(h)
h = self.conv2(g, h)
return h
# Create the model with given dimensions
def train(g, model):
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
best_val_acc = 0
best_test_acc = 0
features = g.ndata['feat']
labels = g.ndata['label']
train_mask = g.ndata['train_mask']
val_mask = g.ndata['val_mask']
test_mask = g.ndata['test_mask']
for e in range(100):
# Forward
logits = model(g, features)
# Compute prediction
pred = logits.argmax(1)
# Compute loss
# Note that you should only compute the losses of the nodes in the training set.
loss = F.cross_entropy(logits[train_mask], labels[train_mask])
# Compute accuracy on training/validation/test
train_acc = (pred[train_mask] == labels[train_mask]).float().mean()
val_acc = (pred[val_mask] == labels[val_mask]).float().mean()
test_acc = (pred[test_mask] == labels[test_mask]).float().mean()
# Save the best validation accuracy and the corresponding test accuracy.
if best_val_acc < val_acc:
best_val_acc = val_acc
best_test_acc = test_acc
# Backward
optimizer.zero_grad()
loss.backward()
optimizer.step()
if e % 5 == 0:
print('In epoch {}, loss: {:.3f}, val acc: {:.3f} (best {:.3f}), test acc: {:.3f} (best {:.3f})'.format(
e, loss, val_acc, best_val_acc, test_acc, best_test_acc))
print(g.ndata['feat'])
print(dataset.num_classes)
model = GCN(g.ndata['feat'].shape[0], 1000, dataset.num_classes).to('cuda')
train(g, model)
I’m getting this error.
Using backend: pytorch
Graph(num_nodes=12651, num_edges=2485,
ndata_schemes={'feat': Scheme(shape=(), dtype=torch.int64), 'label': Scheme(shape=(), dtype=torch.int64), 'train_mask': Scheme(shape=(), dtype=torch.bool), 'val_mask': Scheme(shape=(), dtype=torch.bool), 'test_mask': Scheme(shape=(), dtype=torch.bool)}
edata_schemes={'edges': Scheme(shape=(), dtype=torch.int64)})
Number of categories: 52
Node features
{'feat': tensor([ 18154, 0, 835243411, ..., 17362, 11490,
11490], device='cuda:0'), 'label': tensor([23, 42, 22, ..., 42, 25, 15], device='cuda:0'), 'train_mask': tensor([ True, True, True, ..., False, False, False], device='cuda:0'), 'val_mask': tensor([False, False, False, ..., False, False, False], device='cuda:0'), 'test_mask': tensor([False, False, False, ..., True, True, True], device='cuda:0')}
Edge features
{'edges': tensor([29, 13, 14, ..., 0, 0, 0], device='cuda:0')}
tensor([ 18154, 0, 835243411, ..., 17362, 11490,
11490], device='cuda:0')
52
Traceback (most recent call last):
File "/home/irfan/PycharmProjects/Graphs/two_layers.py", line 84, in <module>
train(g, model)
File "/home/irfan/PycharmProjects/Graphs/two_layers.py", line 51, in train
logits = model(g, features)
File "/home/irfan/environments/Graphs/lib/python3.6/site-packages/torch/nn/modules/module.py", line 727, in _call_impl
result = self.forward(*input, **kwargs)
File "/home/irfan/PycharmProjects/Graphs/two_layers.py", line 32, in forward
h = self.conv1(g, in_feat)
File "/home/irfan/environments/Graphs/lib/python3.6/site-packages/torch/nn/modules/module.py", line 727, in _call_impl
result = self.forward(*input, **kwargs)
File "/home/irfan/environments/Graphs/lib/python3.6/site-packages/dgl/nn/pytorch/conv/graphconv.py", line 417, in forward
graph.srcdata['h'] = feat_src
File "/home/irfan/environments/Graphs/lib/python3.6/site-packages/dgl/view.py", line 81, in __setitem__
self._graph._set_n_repr(self._ntid, self._nodes, {key : val})
File "/home/irfan/environments/Graphs/lib/python3.6/site-packages/dgl/heterograph.py", line 3993, in _set_n_repr
' Got %d and %d instead.' % (nfeats, num_nodes))
dgl._ffi.base.DGLError: Expect number of features to match number of nodes (len(u)). Got 1000 and 12651 instead.