I have implemented GraphSage node classification to classify 4 class, and I’m now trying to change the model from classification to regression since each node has a ground truth value.
What I have done is to change the out_dim to 1, and change the loss function to MAE.
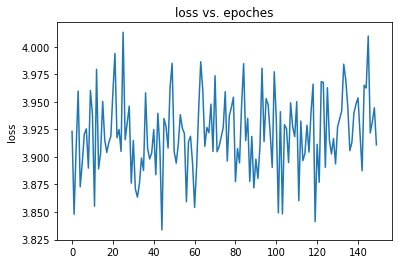
The problem is training loss cannot decrease, I wonder if something wrong about my model.
Here is the model and loss function:
###############Building_model###############
from dgl.nn.pytorch import conv as dgl_conv
####take node features as input and computes node embedding as output
class GraphSAGEModel(nn.Module):
def __init__(self,
in_feats,
n_hidden,
out_dim,
n_layers,
activation,
dropout,
aggregator_type):
super(GraphSAGEModel, self).__init__()
self.layers = nn.ModuleList()
# input layer
self.layers.append(dgl_conv.SAGEConv(in_feats, n_hidden, aggregator_type,
feat_drop=dropout, activation=activation))
# hidden layers
for i in range(n_layers - 1):
self.layers.append(dgl_conv.SAGEConv(n_hidden, n_hidden, aggregator_type,
feat_drop=dropout, activation=activation))
# output layer
self.layers.append(dgl_conv.SAGEConv(n_hidden, out_dim, aggregator_type,
feat_drop=dropout, activation=None))
def forward(self, g, features):
h = features
for layer in self.layers:
h = layer(g, h)
return h
`regression = GraphSAGEModel(feat_num,64,1,2,F.relu,0.5,'gcn')`
for epoch in range(150):
loss_list = []
for batch in range(graph_num):
#model.train()
#logits = model(G,input_list[batch])
#loss_fcn = torch.nn.CrossEntropyLoss()
#loss = loss_fcn(logits[labelnodes_list[batch]], labels_list[batch])
# change loss function
regression.train()
logits = regression(G,input_list[batch])
loss_mae = nn.L1Loss()
labels_list[batch].resize_(len(labels_list[batch]),1) #make tensor has same size [13,1]
loss = loss_mae(logits[labelnodes_list[batch]], labels_list[batch])
optimizer.zero_grad()
loss.backward()
optimizer.step()
loss_list.append(loss.item())
loss_data = np.array(loss_list).mean()
print("Epoch {:05d} | Loss: {:.4f}".format(epoch + 1, loss_data))