Hi,
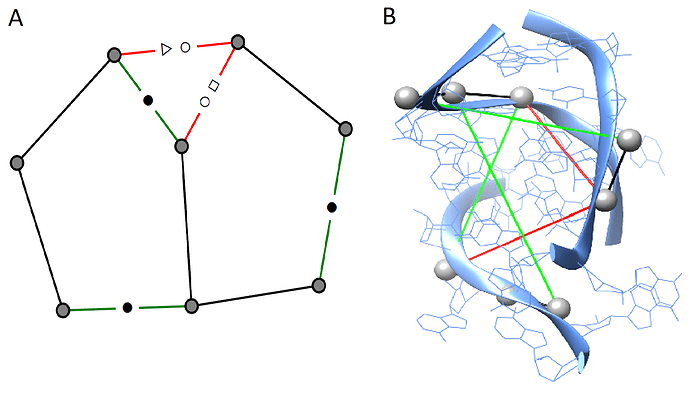
I recently curated a set of RNA from the protein databank into augmented base pairing networks. RNA are biological molecules like proteins. The challenge is to make predictions on which nodes occur at interfaces (regions where the molecule interacts with other proteins, RNA, ions). A useful application in drug discovery.
The graphs I curated distinguish between edge types by their chemical bonding behaviour to record their 3D structure. There are 13 types of edges. The nodes are then labelled by what type of interface they occur in: RNA-RNA, RNA-Protein, RNA-Small-molecule, RNA-Ion. So the task is to use the edge type geometry to try and make a prediction of which nodes occur in an interface. Here is an example of what they look like:
To predict the interface nodes our implementation uses a DGL RGCN to mine recurring structural elements in the graphs (motifs). We made a program for this called vernal
Repo with info on how the data was generated
I was hoping to contribute this dataset to the DGL community to see what learning applications other people can come up with to predict which nodes occur at the interface. How can I contribute to DGL with this topic?