Hello,
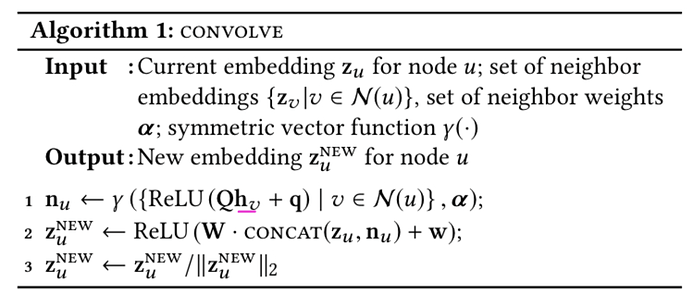
As previously mentioned in the README of the PinSage implementation, there are several deviations between the pseudocode of the original paper and the methodology used in the implementation. If possible, can you confirm whether the differences I will list below are correct?
For simplicity, I am posting the message passing implementation code here (so that I can refer to lines):
1 def forward(g, h, weights):
2 h_src, h_dst = h
3 with g.local_scope():
4 g.srcdata['n'] = self.act(self.Q(self.dropout(h_src)))
5 g.edata['w'] = weights.float()
6 g.update_all(fn.u_mul_e('n', 'w', 'm'), fn.sum('m', 'n'))
7 g.update_all(fn.copy_e('w', 'm'), fn.sum('m', 'ws'))
8 n = g.dstdata['n']
9 ws = g.dstdata['ws'].unsqueeze(1).clamp(min=1)
10 z = self.act(self.W(self.dropout(torch.cat([n / ws, h_dst], 1))))
11 z_norm = z.norm(2, 1, keepdim=True)
12 z_norm = torch.where(z_norm == 0, torch.tensor(1.).to(z_norm), z_norm)
13 z = z / z_norm
-
Dropout: it seems that original paper does not list dropout while the implementation does.
-
Linear projection: it seems that the original paper uses pooling rather than a linear projection to get the hidden representations of neighborhood nodes
-
Normalization by edge sums: it seems that in the implementation, line 10 normalizes the neighbourhood representation by the sum of outgoing edges, this does not seem to be referenced in the original paper.
Please let me know if I have missed something here or if I am somehow incorrect in my understanding.
Thank you in advance!