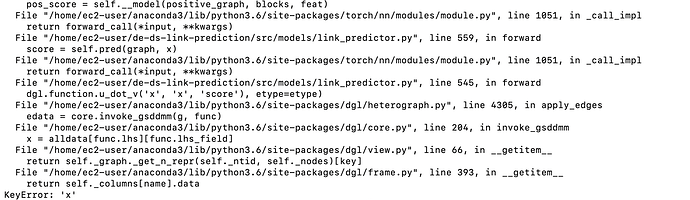
I am training heterogenous link_prediction. My dataset includes transaction connection(edges) between customer and different types of service_provider nodes. So the direction of edges are most one directional but there are some transactions between customer nodes as well. I also added inverse edges( from service_provider to customer for every actual edges) to make it bidirectional for every blocks whikle training as I have seen in one of the discussions link here that only one directional edges in a block can result to this error while training. But still while training my model I face KeyError: 'x' randomly. Sometimes the training would complete without this problem at all. So what is the underlying problem and why is not showing every time I train. Here is the ss of the error message.
It will be great if you could paste your model definition code for a further investigation. From the log you pasted here, it seems that the error happens when it calls into dgl.function.u_dot_v('x', 'x', 'score'), which tries to get feature 'x' from both the source and destination nodes. The KeyError happens when there is no such a feature for either source or destination nodes. My experience is that it is often due to sampling. I think your issue is similar to KeyError: 'x' in GNN for Link Prediction with Neighborhood Sampling - #3 by ogggcar. Please take a look.
This is the model definition code snippet:
class StochasticTwoLayerRGCN(nn.Module):
def __init__(self, in_feat, hidden_feat, out_feat, rel_names):
super().__init__()
self.conv1 = dglnn.HeteroGraphConv({
rel : dglnn.GraphConv(in_feat, hidden_feat, norm='right')
for rel in rel_names
})
self.conv2 = dglnn.HeteroGraphConv({
rel : dglnn.GraphConv(hidden_feat, out_feat, norm='right')
for rel in rel_names
})
def forward(self, blocks, x):
print("inside StochasticTwoLayerRGCN.forward!!!")
x = self.conv1(blocks[0], x)
x = self.conv2(blocks[1], x)
return x
class HeterogeneousScorePredictor(nn.Module):
def forward(self, edge_subgraph, x):
with edge_subgraph.local_scope():
edge_subgraph.ndata['x'] = x
for etype in edge_subgraph.canonical_etypes:
edge_subgraph.apply_edges(
dgl.function.u_dot_v('x', 'x', 'score'), etype=etype)
return edge_subgraph.edata['score']
class HeterogeneousModel(nn.Module):
def __init__(self, in_features, hidden_features, out_features, etypes):
super().__init__()
self.rgcn = StochasticTwoLayerRGCN(
in_features, hidden_features, out_features, etypes)
self.pred = HeterogeneousScorePredictor()
def forward(self, graph, blocks, x):
x = self.rgcn(blocks, x)
score = self.pred(graph, x)
#pos_score = self.pred(positive_graph, x)
#neg_score = self.pred(negative_graph, x)
return score #pos_score, neg_score
I used various print statements to find where the problem is happening. It seems after convolution 1 or more type of connection is lost somehow from the dictionary. Which results key error when assigning node_data to a certain node_type unique to that edge type.
And yes, the link you provided seems to have discussion on similar issue but it doesn’t have solution yet…as it is an active discussion.
This is expected due to the nature of neighbor sampling. Suppose your graph has the following relations:
('user', 'like', 'movie')
('user', 'follow', 'user')
('movie', 'directed-by', 'director')
Suppose you sample from a couple of 'user' nodes. In the first hop, edges of relation ('user', 'like', 'movie') and ('user', 'follow', 'user') could be sampled and some 'movie' and 'user' nodes become the new starting points. Only after the second hop, ('movie', 'directed-by', 'director') will be added and 'director' typed nodes will be needed. Therefore, after the first convolution (which uses the second-hop subgraph), some node/edge types may disappear because they are not needed anymore to compute the next layer.
you are spot-on…but I also added reverse edges for example ('movie', 'liked_by','user') with similar edge data to try and avoid this situation. But it does not seem to work…Is there any way around for this problem
Could you list all the canonical edge types in your graph?
sorry for the late reply…
'PAYMENT': ('CUSTOMER', 'PAYMENT', 'MERCHANT'),
'CI': ('CUSTOMER', 'CI', 'AGENT'),
'CO' : ('CUSTOMER', 'CO', 'AGENT'),
'BILL_PAY' : ('CUSTOMER','BP','BILLER'),
'P2P': ('CUSTOMER', 'P2P', 'CUSTOMER'),
'MR': ('CUSTOMER', 'MR', 'CUSTOMER'),
'P_BACK':('MERCHANT', 'P_BACK', 'CUSTOMER'),
'MR_BACK':('CUSTOMER', 'MR_BACK', 'CUSTOMER'),
'BP_BACK': ('BILLER', 'BP_BACK', 'CUSTOMER'),
'P2P_BACK': ('CUSTOMER', 'P2P_BACK', 'CUSTOMER'),
'CI_BACK': ('AGENT', 'CI_BACK', 'CUSTOMER'),
'CO_BACK':('AGENT', 'CO_BACK', 'CUSTOMER'),
these are the canonical edge types I am using for the experiment
I suspect this is related to sampling, which might be the reason why the error occurred non-deterministically.
I also tried to debug on my on…it seemed that either blocks[0] or blocks[1] has 0 edges of a specific edge_type…which results in this error…now I am using a condition to check whether it is the case before training…and skip if that is the case…
Is this the right approach?
Hi @tanvirbKash , it is difficult for us to debug without a runnable script. Could you please post the entire runnable script here?
class HeterogeneousCreateDataloader:
def __init__(self,graph,inference_graph):
self.__graph = graph
self.__inference_graph = inference_graph
def create_dataloader(self):
data_projection = HeterogeneousDataProjection(self.__graph,self.__inference_graph)
self.__graph = data_projection.train_graph_node_edge_projection()
train_dataloader = self.train_dataloader()
self.__inference_graph = data_projection.test_graph_node_edge_projection()
test_dataloader = self.test_dataloader()
return self.__graph,train_dataloader,self.__inference_graph,test_dataloader
def train_dataloader(self):
train_eid_dict = {}
train_nid_dict = {}
for etype in self.__graph.etypes:
train_eid_dict.update({etype : self.__graph.edges(etype = etype, form = 'eid').squeeze().tolist()})
for ntype in self.__graph.ntypes:
train_nid_dict.update({ntype : self.__graph.nodes(ntype = ntype).squeeze().tolist()})
sampler = dgl.dataloading.MultiLayerNeighborSampler([4, 4])
dataloader = dgl.dataloading.EdgeDataLoader(
self.__graph,
train_eid_dict,
sampler,
negative_sampler=dgl.dataloading.negative_sampler.Uniform(5),
batch_size=BATCH_SIZE,
shuffle=SHUFFLE,
drop_last=DROP_LAST,
num_workers=HETEROGENEOUS_NUM_WORKERS
)
return dataloader
def test_dataloader(self):
test_eid_dict = {}
test_nid_dict = {}
for etype in self.__inference_graph.etypes:
#print(etype)
#print(g.edges(etype = etype, form = 'eid').squeeze().tolist())
test_eid_dict.update({etype : self.__inference_graph.edges(etype = etype, form = 'eid').squeeze().tolist()})
for ntype in self.__inference_graph.ntypes:
test_nid_dict.update({ntype : self.__inference_graph.nodes(ntype = ntype).squeeze().tolist()})
test_sampler = dgl.dataloading.MultiLayerNeighborSampler([4, 4])
test_dataloader = dgl.dataloading.EdgeDataLoader(
self.__inference_graph,
test_eid_dict,
test_sampler,
device=DEVICE, # Put the MFGs on CPU or GPU
g_sampling=self.__graph, # Graph to sample neighbors
#negative_sampler=dgl.dataloading.negative_sampler.Uniform(5),
batch_size=BATCH_SIZE,
shuffle=SHUFFLE,
drop_last=DROP_LAST,
num_workers=HETEROGENEOUS_NUM_WORKERS
)
return test_dataloader
class StochasticTwoLayerRGCN(nn.Module):
def __init__(self, in_feat, hidden_feat, out_feat, rel_names):
super().__init__()
self.conv1 = dglnn.HeteroGraphConv({
rel : dglnn.GraphConv(in_feat, hidden_feat, norm='right')
for rel in rel_names
})
self.conv2 = dglnn.HeteroGraphConv({
rel : dglnn.GraphConv(hidden_feat, out_feat, norm='right')
for rel in rel_names
})
def forward(self, blocks, x):
print("inside StochasticTwoLayerRGCN.forward!!!")
x = self.conv1(blocks[0], x)
print(blocks[0])
print(x.keys())
print(blocks[1])
x = self.conv2(blocks[1], x)
return x
class HeterogeneousScorePredictor(nn.Module):
def forward(self, edge_subgraph, x):
with edge_subgraph.local_scope():
#print("assigning x")
edge_subgraph.ndata['x'] = x
print("New edge_subgraph!")
print(edge_subgraph.ndata['x'].keys())
for etype in edge_subgraph.canonical_etypes:
print(etype)
#print(x[etype].shape)
edge_subgraph.apply_edges(
dgl.function.u_dot_v('x', 'x', 'score'), etype=etype)
return edge_subgraph.edata['score']
class HeterogeneousModel(nn.Module):
def __init__(self, in_features, hidden_features, out_features, etypes):
super().__init__()
self.rgcn = StochasticTwoLayerRGCN(
in_features, hidden_features, out_features, etypes)
self.pred = HeterogeneousScorePredictor()
def forward(self, graph, blocks, x):
print("Inside Model.forward()")
print(x.keys())
x = self.rgcn(blocks, x)
print(x.keys())
score = self.pred(graph, x)
#pos_score = self.pred(positive_graph, x)
#neg_score = self.pred(negative_graph, x)
return score #pos_score, neg_score
class HeterogeneousLinkPredictor:
def __init__(self,train_graph, train_dataloader,test_graph,test_dataloader,edge_weight): #will use test_dataloader later
self.__train_dataloader = train_dataloader
self.__train_graph = train_graph
self.__test_dataloader = test_dataloader
self.__test_graph = test_graph
self.__train_edge_weight = edge_weight
print("HeterogeneousLinkPredictor.init()")
print(self.__train_graph.etypes)
self.__model = HeterogeneousModel(HETERO_IN_FEAT, HETERO_HIDDEN_FEAT, HETERO_OUT_FEAT, self.__train_graph.etypes)
#self.__predictor = HeterogeneousScorePredictor().to(DEVICE)
self.__opt = torch.optim.Adam(self.__model.parameters())
def training(self):
skip_flag = 0
#model = Model(in_features, hidden_features, out_features, etypes)
#model = model.cuda()
for epoch in range(int(EPOCHS)):
with tqdm.tqdm(self.__train_dataloader) as tq:
for step, (input_nodes, positive_graph, negative_graph, blocks) in enumerate(tq):
#positive_graph = positive_graph.to(torch.device('cpu'))
#negative_graph = negative_graph.to(torch.device('cpu'))
feat = blocks[0].srcdata['feat'] #blocks[0].srcdata['features']
print("Inside Training")
print(feat.keys())
if blocks[0].num_nodes('MERCHANT') == 0 or blocks[0].num_nodes('CUSTOMER') == 0 or blocks[0].num_nodes('BILLER') == 0 or blocks[0].num_nodes('AGENT') == 0 or blocks[1].num_nodes('MERCHANT') == 0 or blocks[1].num_nodes('CUSTOMER') == 0 or blocks[1].num_nodes('BILLER') == 0 or blocks[1].num_nodes('AGENT') == 0:
print("H1")
print(blocks[0])
print(blocks[1])
epoch-=1
skip_flag+=1
continue
pos_score = self.__model(positive_graph, blocks, feat)
neg_score = self.__model(negative_graph, blocks, feat)
### need to scale and sum the loss...
total_loss = 0
for p_key,n_key in zip(pos_score.keys(),neg_score.keys()):
#print(pos_score[p_key].shape[0])
if pos_score[p_key].shape[0] == 0:
#print("H2")
continue
loss = compute_loss(pos_score[p_key], neg_score[n_key])
total_loss+= self.__train_edge_weight[p_key]*loss
#torch.detach_()
tq.set_postfix({'loss': '%.03f' % total_loss.item()}, refresh=False)
self.__opt.zero_grad()
total_loss.backward()
self.__opt.step()
auc = {}
thresholds = {}
with torch.no_grad():
input_nodes, positive_graph, negative_graph, blocks = next(iter(self.__train_dataloader))
feat = blocks[0].srcdata['feat']
pos_score = self.__model(positive_graph, blocks, feat)
neg_score = self.__model(positive_graph, blocks, feat)
for p_key,n_key in zip(pos_score.keys(),neg_score.keys()):
auc.update(
{p_key:compute_auc(pos_score[p_key], neg_score[n_key])}
)
for p_key,n_key in zip(pos_score.keys(),neg_score.keys()):
fpr,tpr,threshold = compute_roc(pos_score[p_key], neg_score[n_key])
# Calculate Youden's J-Statistic to identify the best threshhold
j_stat = tpr - fpr
# locate the index of the largest J
ix = np.argmax(j_stat)
#print(threshold)
best_thresh = threshold[ix]
#print('Best Threshold: %f' % (best_thresh))
thresholds.update(
{p_key:best_thresh}
)
print(skip_flag)
return auc,thresholds
Used many print statements in different places for debug purposes…
Big if statement in def testing() is to avoid the KeyError…wondering if that is alright or not
Thanks for the code. I may need some time before coming back to this. Please ping me if there is no update next week.
This topic was automatically closed 30 days after the last reply. New replies are no longer allowed.