I was checking the code and realized that the node features are indeed aggregated at each iteration.
I have a graph classification task for graphs containing 4 nodes (3 features per node) and 3 edges (1 edge feature). However, I couldn’t find the line of code that considers g.edata so I was wondering whether the source code applies to my problem as the MiniGCDataset does not contain node and edge features. Is there something specific I should add to account for edge features ?
We have some model examples that use edge features for batched graph regression/classification, such as mpnn and attentive fp.
@mufeili Thanks a lot ! I have tried mpnn for my regression purposes. I just have one last question to check whether GCN is suitable for me:
I have 300 000 graph samples, with each graph having 4 nodes , 3 features per node and 3 edges connecting them. There are 4 classes in total for all the graphs.I have trained them using the source code for Batched graph classification, but the loss is not decreasing significantly and the evaluation shows all graphs recognized as class 1 (although i made sure to balance my data).
Is my graph too sparse for GCN with only 4 nodes ? Or can the problem be elsewhere ?
Any advice would be appreciated 
EDIT: I just printed the output of my model on the testing batch and it seems the torc.softmax output is the same for all entries, so there must be something wrong in the model definition for my case! (i used the same Classifier) model_test|489x180
Hi, I agree this is weird. Here are some follow-up questions:
- Did you try some baseline models like MLPs? How did they perform?
- What do you mean by “3 edges connecting them”? Do you mean each graph has 3 edges in total?
- For the graphs you described, it seems that the topologies of them will be quite similar so I assume node features will be quite different for each class?
- MPNN might be an overkill for simple graphs like this, I will start with GCN and get it to work first.
@mufeili Thank you for your reply. To better explain my problem :
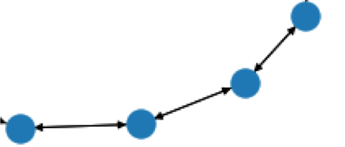
One graph consists simply of 4 nodes as shown in the attached photo below. Each node has 3 continuous features ranging from -10 to 10. I have a total of 400 000 of such graphs for training. Each node has various continuous node features ranging from -10 to 10. My trials using conv2d and MLPs on these continuous data (treated as 400 000 samples of 4x3 arrays) were successful. Now I am using the batched graph classification to classify each of these graphs (there are 4 classes), but the loss is not decreasing, even when I change the learning rate, add more GCN layers and try different loss functions. I also modeled tried both directed and un-directed approaches. Any advice would be appreciated 
Hi @aah71, your graphs look like chains. There have been some study that GNNs may not work well on highly regular graphs like chains. See How powerful are Graph Convolutions?. How are these graphs constructed? If there is not an important semantic meaning associated with the graph topology, I don’t think using GNNs will be a very good idea in this case.
So glad I saw this! The mpnn model rocks. You just pass in like so
nodes_weights = g.ndata['n_weight']
edges_weights = g.edata['e_weight']
prediction = model(g, nodes_weights, edges_weights)
In my dream dgl world, you could pass either ndata and/ or edata in as params into any model. From there, a model-agnostic function would make tensors for each key of the appropriate dimensions.